Abstract
Mitochondrial complete genome (mtDNA) of Hoplias intermedius, commonly known as “traírão” is presented. DNA from a muscle tissue sample was sequenced by next-generation sequencing. To corroborate the results, phylogenetic analysis was performed with MEGA6 to compare the complete mitogenome of H. intermedius with genomes of other Characiformes species. The mtDNA molecule was circular and 16,629 bp in length. Gene content included 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes and a D-loop noncoding region. All genes had ATG as the start codon. Six genes carried complete stop codons of AGG (COI), TGA (ND6) and TAA (ND1, ATPase 8, ND4L and ND5), while seven genes had incomplete stop codons. Phylogenetic relationships with other Ostariophysi species placed H. intermedius together with H. malabaricus as a monophyletic group belonging to the order Characiformes.
Genus Hoplias has a widespread distribution across South American rivers. Two species are currently represented in the São Francisco river basin: Hoplias intermedius and Hoplias malabaricus (Reis et al. Citation2003). The geographic distribution of H. intermedius comprises the headwaters of Paraná river basin (Oyakawa & Mattox Citation2009), in contrast to H. malabaricus that has a widespread distribution across South and Central America (Reis et al. 2003). Taxonomic characters differentiates H. intermedius from H. malabaricus by the number of scales on lateral sensorial channel between 42 and 46 and by the contralateral dentary margins running parallel to each other in H. intermedius (vs. dentary margins with a triangular shape in H. malabaricus) (Oyakawa & Mattox Citation2009). Owing to its importance as a fishery resource, its restricted distribution and the negative impacts of mining activities, H. intermedius was selected as the subject of this study (Alves & Pompeu Citation2010; Vieira et al. Citation2005).
Taxonomic mislabeling represents an obstacle in forensic analysis. In this context, the characterization of mDNA, considering its well-conserved sequence, high abundance and small circular structure (Bilington & Hebert Citation1991; Pimentel et al. Citation2014), represents a useful information source for species identification.
Specimens were collected at Cipó river at Minas Gerais State, Brazil (19°20′01.1″S, 43°39′17.0″W) and stored in the UFMG tissue collection (UFMG-BDT-PP000004). Total genomic DNA was extracted from muscle samples by the method described by Herrmann and Frischauf (Citation1987). Genomic library was constructed by using a paired-end 600-cycle strategy with the MiSeq platform (Illumina Inc., San Diego, CA).
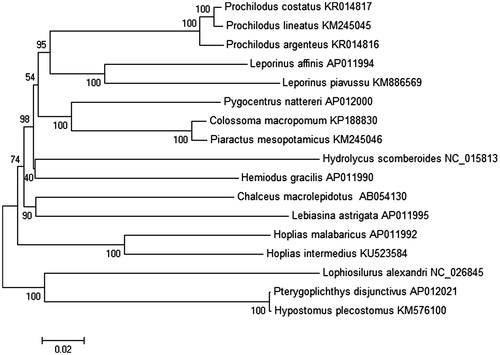
Quality control of generated reads and sequence assembly were performed using CLC Workbench software v8.5.1 (CLC Bio-Qiagen, Aarhus, Denmark). Contigs were annotated by using the MitoFish webserver (Iwasaki et al. Citation2013). De novo assembly was solved with 304.45% coverage (GenBank accession number: KU523584). The annotated genome contained 35 coding and one noncoding region. To compare the mitogenome characteristics among Characiformes, the complete mitochondrial genome of all 15 neotropical species available in the GenBank database were retrieved and used for phylogenetic analysis in Mega6 (Tamura et al. Citation2013) (), applying neighbour-joining algorithm.
Figure 1. Neighbour-joining distance consensus tree based on complete mitochondrial DNA (without D-loop noncoding region), explaining a hypothesis of phylogenetic relationships among seven families from order Characiformes and two families from order Siluriformes as the outgroup. Members of family Erythrinidae (H. intermedius, H. malabaricus) are grouped in a single clade belonging to order Characiformes, showing a high sequence similarity. The Prochilodontidae clade (Prochilodus argenteus (P. costatus, P. lineatus)) confirms the results obtained by Chagas et al. (Citation2016). The phylogenetic tree was constructed under the Kimura-2 parameter model and consensus tree using 1000 bootstrap. Numbers indicate support of each clade, showing higher supports for relationships between species of the same family and lower supports of relationships grouping different families between order Characiformes.
The mtDNA of H. intermedius is a circular DNA molecule of 16,629 bp, 43.96% of GC content. Start codon ATG was found in 13 protein-coding genes encoded on the heavy strand, and as expected, in ND6 encoded on the light strand. Four PCGs (ND1, ATPase8, ND4L, ND5) contained the complete TAA stop codon, similar to Salminus brasiliensis (Brandão-Dias et al. Citation2016) and genus Prochilodus (Carmo et al. Citation2016; Chagas et al. Citation2016). One gene (COI) contained AGG as the stop codon, similar to Brycon orbignyanus (Siqueira et al. Citation2014) and Piaractus mesopotamicus (Pimentel et al. Citation2016). One gene (ND6) contained TGA as the stop codon, in contrast with other Bryconidae, Prochilodontidae and Serrasalmidae species. Five genes (ND2, ND3, ND4, COII, Cytb) contained incomplete T–– stop codons, and two genes (ATPase 6, COIII) displayed incomplete TA– stop codons. Incomplete stop codons were probably completed as TAA by posttranscriptional polyadenylation (Ojala et al. Citation1981).
Funding information
Financial support was provided by CAPES (Edital Ci^encias Forenses no.25/2014) and CEMIG project PD455.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Alves CBM, Pompeu PS. 2010. Peixes do Rio das Velhas: passado e presente. Belo Horizonte, Brazil: SEGRAC.
- Bilington N, Hebert P. 1991. Mitochondrial DNA diversity in fishes and its implications for introduction. Can J Fish Aquat Sci. 48:80–94.
- Brandão-Dias PFP, Carmo AO do, Martins APV, Pimenta RJG, Alves CBM, Kalapothakis E. 2016. Complete mitochondrial genome of Salminus brasiliensis. Mitochondrial DNA. 27:1577–1578.
- Carmo AO, Brandão-Dias PFP, Martins APV, Bedore AG, Kalapothakis E. 2016. Complete mitochondrial genome sequence of Prochilodus lineatus (Characiformes, Prochilodontidae). Mitochondrial DNA. 27:1946–1947.
- Chagas ATA, Carmo AO, Costa MA, Resende LC, Brandao-Dias PFP, Martins APV, Kalapothakis E. 2016. Description and comparison of two economically important fish species mitogenomes: Prochilodus argenteus and Prochilodus costatus (Characiformes, Prochilodontidae). Mitochondrial DNA. 27:2852–2853.
- Herrmann BG, Frischauf AM. 1987. Isolation of genomic DNA. Meth Enzymol. 152:180–183.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, Nishida M. 2013. MitoFish and Mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 11:2531–2540.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Oyakawa OT, Mattox GMT. 2009. Revision of the Neotropical trahiras of the Hoplias lacerdae species-group (Ostariophysi: Characiformes: Erythrinidae) with descriptions of two new species. Neotrop Ichthyol. 7:117–140.
- Pimentel JSM, Carmo AO, Maciel DL, Siqueira FF, Kalapothakis E. 2016. Complete mitochondrial genome sequence of Piaractus mesopotamicus (Holmberg, 1887). Mitochondrial DNA. 27:1940–1941.
- Reis RE, Kullander SO, Ferraris CJJ. 2003. Check List of the Freshwater Fishes of South and Central America (CLOFFSCA). 1st ed. Porto Alegre: EDIPUCRS.
- Siqueira FF, Carmo AO, Pimentel JSM, Kalapothakis E. 2014. Complete mitochondrial sgenome sequence of Brycon orbignyanus (Characiformes, Bryconidae). Mitochondrial DNA. 27:1942–1943.
- Vieira F, Santos GB, Alves CBM. 2005. A ictiofauna do Parque Nacional da Serra do Cipó (Minas Gerais, Brasil) e áreas adjacentes. Lundiana. 6:77–87.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
