Abstract
Here we report the complete mitochondrial genome of the emperor dragonfly, Anax imperator (Odonata: Aeshnidae) as the first of its genus. Data were generated via next generation sequencing (NGS) and assembled using an iterative approach. The typical metazoan set of 37 genes (13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes) was detected in the same gene order as in other odonate mitogenomes. However, only three intergenic spacer regions are present in A. imperator lacking the distinct s5 spacer, which was regarded as informative feature of the odonate suborder Anisoptera (dragonflies) but absent in Zygoptera (damselflies). With 16,087 bp, it is the longest anisopteran mitogenome to date, mainly due to the long A + T-rich control region of 1291 bp.
The emperor dragonfly, Anax imperator, is a widespread and common species in the old world inhabiting all types of standing- and slow-running freshwater ecosystems. It was one of the first odonate species for which a recent range shift northwards (e.g. Parr Citation2010) and towards higher altitudes (Westermann Citation2003; Hunger et al. Citation2006) was noticed due to global climate change. The first records of this species in Sweden were 2002 (Ott Citation2010). In only 11 years A. imperator crossed a distance of 970 km northwards through Scandinavia (Nielsen Citation1998; Lejfelt-Sahlén Citation2007). The larvae of this large dragonfly species are known to be very aggressive (e.g. Beutler Citation1985) and will invade and significantly influence the native species composition of freshwater ecosystems. Genetic and comparative genomic studies on range shift, expansion, and adaptive potential of this species are of great interest to further elucidate the impact of global change on flying insects. To date for A. imperator, a panel of 10 nuclear microsatellite loci and partial mitochondrial genes (cox1, nad1, and both rRNAs) were established so far to serve in various phylogenetic studies (Misof et al. Citation2001; Hadrys et al. Citation2007; Fleck et al. Citation2008; Rach et al. Citation2008; Bergmann et al. Citation2013). To consequently proceed towards a comparative genomic approach one first step is the unravelling and comparison of mitogenomes, e.g. their gene content, arrangements, and genealogical relationships.
As for the A. imperator mitogenome, a standard phenol–chloroform protocol by Hadrys et al. (Citation1992) was used to extract total genomic DNA from flight muscles of a single individual collected in Southern France (43°36′17.7″N 4°48′34.4″E). DNA was submitted for library preparation and whole genome sequencing on an llumina HiSeq2000 (75 bp paired-end reads) to the Yale Center for Genome Analyses (YCGA, http://www.ycga.yale.edu). Different mitochondrial gene sequences containing partial nad1, cox1, 12S rRNA, and 16S rRNA genes (accession numbers: KC912228.1, KF584974.1, EU477652.1 and EU183256.1) were used as reference seeds for a subsequent assembly employing Genious v.8.1.5 (http://www.geneious.com/). For mitochondrial genome annotation, the MITOS WebServer (mitos.bioinf.uni-leipzig.de/index.py) was applied and results were checked manually using BLAST (Altschul et al. Citation1990) and available odonate mitochondrial genomes (e.g. Yu et al. Citation2014; Chen et al. Citation2015). Transfer RNA genes were predicted using both, the tRNAscan-SE v.1.21 Search Server (Lowe & Eddy Citation1997) and ARWEN v.1.2 (Laslett & Canbäck Citation2008).
The complete circular mitochondrial genome sequence of A. imperator (GenBank accession number #KX161814) with the length of 16,087 bp is the largest known mitogenome among Anisoptera. It exhibits the standard metazoan gene content of 37 genes, comprising 13 protein-coding genes, 22 tRNA genes, and two rRNA genes which are identically arranged as in the few other odonate mitochondrial genomes (e.g. Simon & Hadrys Citation2013; Lorenzo-Carballa et al. Citation2014; Chen et al. Citation2015; Yu et al. Citation2014; Feindt et al. Citation2016). Overall base frequency is 76.0% AT-biased, for the 1291 bp long control (A + T rich) region even 93.5%. All standard mitochondrial invertebrate start codons are found, in detail ATT (nad5), ATA (nad2, nad3), TTG (cox1, nad1), ATC (atp8, nad6), and ATG (cox2, atp6, cox3, nad4, nad4l, cob). Two proteins (cox2, nad5) possess a single T as an incomplete stop codon, requiring post-transcriptional polyadenylation whereas all others protein-coding genes use TAA as stop codon (). The gene length of tRNA genes ranges from 65 bp to 73 bp and all tRNAs can be folded in the typical cloverleaf structure, except the D-replacement tRNA trnS1. Further, two pseudo-tRNA genes were detected by the tRNA prediction software ARWEN v.1.2 (Laslett & Canbäck, Citation2008) which were both D-Loop tRNAs and located inside the cox2 sequence and in trnA/trnR, respectively. Therefore, their functionality remains questionable.
Table 1. Mitochondrial genome organization and gene content of A. imperator with detailed description of gene boundaries, strand, gene length (in bp) as well as start and stop codons for protein-coding genes and anticodons for tRNA genes, respectively.
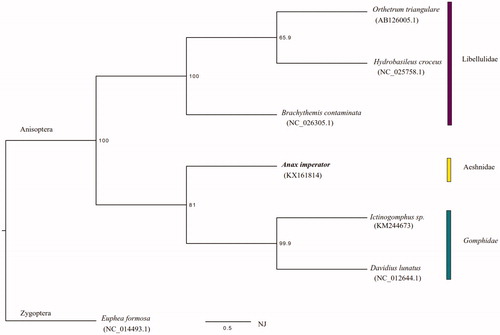
However, in contrast to the known other anisopteran mitogenomes, only three intergenic spacer regions were discovered (see ). These are located between trnY/cox1, trnT/trnP, and trnS2/nad1. They are also present in other odonates (Anisoptera and Zygoptera), e.g. Ischnura elegans (Feindt et al. Citation2016), Ischnura pumilio (Lorenzo-Carballa et al. Citation2014), Megaloprepus caerulatus (Feindt et al. Citation2016), or Brachythemis contaminata (Yu et al. Citation2014). The latter, an anisopteran species additionally shows a fourth spacer region between nad1/trnL2 that is asserted to be typical for Anisopterans and lacking in Zygopterans (Lin et al. Citation2010). This spacer, commonly called s5 (though counting and numbering spacer regions is not consistent between most mitogenome publications) is not present in Anax. Consequently, the absence of this spacer refutes the theory of being a putative distinctive feature between Anisoptera and Zygoptera and stresses the necessity to analyze more mitogenomes within Odonata to allow stronger, reliable assumptions about phylogenetically informative mtDNA characteristics. The phylogenetic position of A. imperator in the context of all available anisopteran mitogenomes to date (3 May 2016) is displayed in and so far consistent with other gene tree phylogenies.
Figure 1. Neighbour-Joining Tree of A. imperator within all available anisopteran odonate species (03 May 2016): Orthetrum triangulare (AB126005.1), Hydrobasileus croceus (NC_025758.1), B. contaminata (NC_026305.1), Ictinogomphus sp. (KM244673) and Davidius lunatus (NC_012644.1). The phylogeny was reconstructed based on 13 mitochondrial protein-coding genes via Paup with 1000 bootsrap replicates and Euphea formosa (NC_014493.1) as an outgroup.
Disclosure statement
The authors declare no conflict of interest to other working groups.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Bergmann T, Rach J, Damm S, DeSalle R, Schierwater B, Hadrys H. 2013. The potential of distance‐based thresholds and character‐based DNA barcoding for defining problematic taxonomic entities by CO1 and ND1. Mol Ecol Resources. 13:1069–1081.
- Beutler H. 1985. [Freiland-Daten zur Koexistenz von Aeshnidenlarven]. Entomol Nachrichten Und Berichte. 29:73–75. [in German].
- Chen M-Y, Chaw S-M, Wang J-F, Villanueva RJT, Nuneza OM, Lin C-P. 2015. Mitochondrial genome of a flashwing demoiselle, Vestalis melania from the Philippine Archipelago. Mitochondrial DNA. 26:720–721.
- Feindt W, Herzog R, Osigus H-J, Hadrys H. 2016. Short read sequencing assembly revealed the complete mitochondrial genome of Ischnura elegans (Vander Linden, 1820). Mitochondrial DNA. 1:547–576.
- Feindt W, Osigus H-J, Herzog R, Mason CE, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the Neotropical helicopter damselfly Megaloprepus caerulatus Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA Part B. 1:497–499.
- Fleck G, Ullrich B, Brenk M, Wallnisch C, Orland M, Bleidissel S, Misof B. 2008. A phylogeny of anisopterous dragonflies (Insecta, Odonata) using mtRNA genes and mixed nucleotide/doublet models. J Zool Syst Evol Res. 46:310–322.
- Hadrys H, Balick M, Schierwater B. 1992. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Molecular Ecol. 1:55–63.
- Hadrys H, Timm J, Streit B, Giere S. 2007. A panel of microsatellite markers to study sperm precedence patterns in the emperor dragonfly Anax imperator (Odonata: Anisoptera). Mol Ecol Notes. 7:296–298.
- Hunger H, Schiel F-J, Kunz B. 2006. [Verbreitung und Phänologie der Libellen BadenWürttembergs (Odonata)]. Libellula Suppl. 7:15–188. [in German].
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lejfelt-Sahlén A. 2007. [Trollsländefauna i förvandling]. Fauna Och Flora. 102:44–44. [in Swedish].
- Lin CP, Chen MY, Huang JP. 2010. The complete mitochondrial genome and phylogenomics of a damselfly, Euphaea formosa support a basal Odonata within the Pterygota. Gene. 468:20–29.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondrial DNA. 25:247–248.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Misof B, Rickert AM, Buckley TR, Fleck G, Sauer KP. 2001. Phylogenetic signal and its decay in mitochondrial SSU and LSU rRNA gene fragments of Anisoptera. Mol Biol Evol. 18:27–37.
- Nielsen OF. 1998. [Guldsmede-nyt fra Danmark 1997]. Nordic Odonatol Soc Newslett. 4:4. [in Danish].
- Ott J. 2010. Dragonflies and climatic change-recent trends in Germany and Europe. BioRisk. 5:253.
- Parr AJ. 2010. Monitoring of Odonata in Britain and possible insights into climate change. In: Ott J (Ed) Monitoring Climatic Change with Dragonflies. BioRisk. 5:127–139.
- Rach J, DeSalle R, Sarkar IN, Schierwater B, Hadrys H. 2008. Character-based DNA barcoding allows discrimination of genera, species and populations in Odonata. Proc Royal Soc London B: Biol Sci. 275:237–247.
- Simon S, Hadrys H. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69:393–403.
- Westermann K. 2003. [Zum Status der Großen Königslibelle im höheren Schwarzwald]. Naturschutz Am Südlichen Oberrhein. 4:81–85. [in German].
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2014. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA. 27:1–2.
