Abstract
Odonata (dragonflies and damselflies) is a small order at the base of flying insects (Pterygota). Resolving family-level phylogenetic relationships within this order receives great attention. Hereby, genetic data already resulted in various changes, which are however still under discussion. Mitochondrial genomes may further enhance such phylogenies. This study presents the complete mitochondrial genome of the Neotropical damselfly Megaloprepus caerulatus based on next generation sequencing (NGS) data on total genomic DNA. The total length comprises 16,094 bp and includes the standard metazoan set of 37 genes together with a 1376 bp long A + T rich (control) region. Gene content, gene arrangement and base frequency are consistent with other odonate mitochondrial genomes. It further contains four intergenic spacer regions, indicating a possible family specific feature for the Coenagrionidae and its close relatives.
The relatively small insect order Odonata owns a key position in the evolution of winged insects and as sensitive indicator organisms for freshwater ecosystems. Robust phylogenies are one ultimate requirement for studies in evolution, ecology and developmental biology. Over the last 10 years many molecular data-based attempts have already resulted in various reorganizations of phylogenetic relationships (e.g. Dumont et al. Citation2010; Carle et al. Citation2015). However, today’s phylogenies still harbour open questions concerning family-level as well as deeper taxonomic positions. Mitochondrial genome projects in odonates may help to unravel such unresolved phylogenetic relationships by constructing more robust phylogenies based on complete gene and genome comparison (e.g. Simon & Hadrys Citation2013). One family that has received great attention in the past are the Pseudostigmatidae (giant damselflies) (e.g. Groeneveld et al. Citation2007; Ingley et al. Citation2012). They were recently placed into the Coenagrionidae based on three sequence markers (Dijkstra et al. Citation2014). Here, more robust genomic data are needed not only to clarify their taxonomic position but also to facilitate genomic-based studies on Neotropical forest health indicators. In this work, we present a complete mitochondrial genome of Megaloprepus caerulatus as the first member of this group. Megaloprepus caerulatus was already included in many ecological (e.g. Fincke & Hedström Citation2008) and evolutionary studies in tropical habitats, but yet little research has been done on its genetics (e.g. Fincke & Hadrys Citation2001; Feindt et al. Citation2014).
Via the standard phenol–chloroform extraction, (Hadrys et al. Citation1992) total genomic DNA from the flight muscles of a single M. caerulatus individual collected at the Biological Research Station La Selva (OTS), Costa Rica (N 10°25′19.74″W 84°00′35.22″) was extracted. Library preparation and DNA sequencing (100 bp mate pairs with different insert sizes, Illumina HiSeq2500, Illumina Inc., San Diego, CA) was performed at the Weill Cornell Medical College in New York. The mitochondrial genome was assembled using Geneious vers. 8.1 (http://www.geneious.com); while mapping a fraction of the cleaned reads onto a seed sequence (here, cox1: KF895301.1 and nad1: KF895193.1) allowing strand extension using varying iterates. Hereby, the settings included a minimum overlap of 60 bp, a minimal overlap identity of 90%, and a variable word size between 35 and 50. The mitochondrial genome was annotated using the MITOS WebServer (mitos.bioinf.uni-leipzig.de/index.py) and verified via BLAST (Altschul et al. Citation1990) against the NCBI database (http://www.ncbi.nih.gov) or additionally with published mitochondrial genomes of other odonate species. Transfer RNA genes were identified by a tRNA covariance model implied on the tRNAscan-SE vers. 1.21 Search Server (http://lowelab.ucsc.edu/tRNAscan-SE; Lowe & Eddy Citation1997) and ARWEN vers. 1.2 (http://mbio-serv2.mbioekol.lu.se/ARWEN, Laslett & Canbäck Citation2008). Phylogenetic relationships were reconstructed using a selection of odonate mitochondrial genomes and a mayfly Parafronurus youi as outgroup (). All 13 protein coding genes and rRNA genes were aligned independently, then concatenated and a maximum parsimony tree was calculated (1000 replicates) in PAUP (Swofford Citation2002).
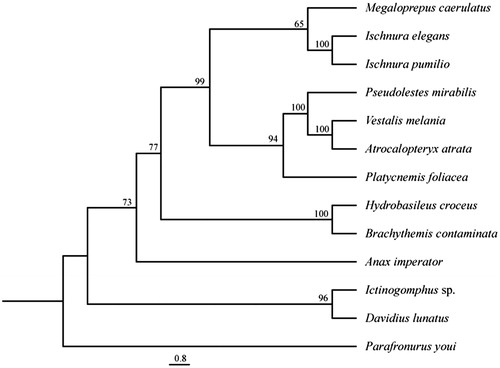
Figure 1. Phylogenetic relationships of odonate species based on maximum parsimony analysis of concatenated mitochondrial protein coding genes and rRNA sequences: Brachythemis contaminata (NC_026305), Ictinogomphus sp. (KM244673), Hydrobasileus croceus (NC_025758), Davidius lunatus (NC_012644), Ischnura pumilio (NC_021617), Pseudolestes mirabilis (NC_020636), Atrocalopteryx atrata (NC_027181), Vestalis melania (NC_023233), Platycnemis foliacea (NC_027180), Anax imperator (KX161841), Ischnura elegans (KU958378), and Parafronurus youi (EU349015.1) as outgroup. The heuristic search (under the 50% majority-rule with 1000 bootstrap replicates) placed Megaloprepus as a sister species to Ischnura spp, whereas relationships are displayed as in other phylogenies.
The obtained complete mitochondrial genome of M. caerulatus (NCBI: KU958377) is the first of a tropical odonate species. It has a total length of 16,094 bp, and is the second largest aside of Vestalis melania (16,685 bp, NC_023233). In all observed parameters, the presented mitochondrial genome shows a strong similarity to all other already published odonate mitochondrial genomes (e.g. Lorenzo-Carballa et al. Citation2014; Tang et al. Citation2014; Yu et al. Citation2014; Chen et al. Citation2015; Feindt et al. Citation2016; Herzog et al. Citation2016). It contains the common arrangement of 37 genes including 13 protein-coding genes, 2 rRNA (16S and 12S rRNA) genes, 22 tRNA genes, and an A + T rich (control) region of 1376 bp in length. Comparing with the previously released single genes, we observed 100% similarity to nad1 (DQ642992.1) (Groeneveld et al. Citation2007) and 16S rRNA (DQ642987.1), and in nad1 (JQ966612.1), 16S (JQ966660.1) and 12S rRNA (JQ966647.1) a similarity between 98% and 100% to genes illustrated in Ingley et al. (Citation2012). The overall A + T content of the mitochondrial genome is 75.9% (A: 43.1%, C: 14.2%, G: 9.8%, T: 32.8%) and therefore it is similar to the base frequencies of the protein-coding genes (AT: 74.4%). Hereby, atp8 encompasses the highest A + T content with 81.1% and cox1, as it was described in other odonates (Lorenzo-Carballa et al. Citation2014), with 68.9% the lowest. All protein coding genes start with characteristic invertebrate specific mitochondrial start codons: cox1, atp6, cox3, nad4, nad4L and cob use ATG; nad2, cox2 and nad6 start with ATA; nad3 and nad1 start with TTG; nad5 starts with ATT and atp8 starts with ATC. The standard stop codon TAA was used eight times (nad2, cox1, atp8, atp6, nad4L, nad6, cob, nad1), whereas TAG was used only once by nad3. An incomplete stop codon with a single T was found in four cases (cox2, cox3, nad5 and nad4). Length of the tRNA genes in M. caerulatus ranges from 64 bp to 74 bp except for trnL1 (Leu), trnS1 (Ser), trnG (Ser) all of which employ the typical clover-leaf secondary structures.
A difference in numbers of intergenic spacer regions is described in the two large odonate orders (Lorenzo-Carballa et al. Citation2014). They seem to provide a phylogenetic signal for the split of Anisoptera (dragonflies) from Zygoptera (damselflies). The lack of the intergenic spacer region s5 seems to be a damselfly specific character. In M. caerulatus, we detected four spacer regions at trnY/cox1, trnF/nad5, trnT/trnP and trnS2/nad1 with a total length of 106 bp. With this we proved that the unique spacer s4 between trnF and nad5, which is also present in I. pumilio (NC_021617), Pseudolestes mirabilis (NC_020636) and I. elegans (KU958378) might not only be a specific character for the Coenagrionidae. The phylogenetic tree () places M. caerulatus as a sister species to Ischnura spp. with low support. More mitochondrial genomic data is needed to resolve relationships within and between families. However, the mitochondrial genome presented in this study is a valuable resource for future population genomic studies and also owns potential for answering phylogenetic questions on higher taxonomic levels.
Funding information
This work was supported by the Annette Kade Graduate Student Fellowship Program of the RGGS at the American Museum of Natural History through generous contributions of the Annette Kade Charitable Trust. We are further grateful for financial support from the German Academic Exchange Service (DAAD) and the Graduate Academy of the Leibniz University Hannover given to WF and a DFG grand (HA1947/6-1) given to HH. HJO acknowledges a doctoral fellowship of the Studienstiftung des deutschen Volkes.
Disclosure statement
Collecting permit was processed from the Ministerio de Ambiente y Energía, Costa Rica (MINAE) and the permit for using the genetic material of the individual was approved by Comisión Nacional para la Gestión de la Biodiversidad, Costa Rica (CONAGEBIO). Furthermore, the authors state no conflicts of interest.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Carle FL, Kjer KM, May ML. 2015. A molecular phylogeny and classification of Anisoptera (Odonata). Arthropod Syst Phylo. 73:281–301.
- Chen M-Y, Chaw S-M, Wang J-F, Villanueva RJT, Nuneza OM, Lin C-P. 2015. Mitochondrial genome of a flashwing demoiselle, Vestalis melania from the Philippine Archipelago. Mitochondrial DNA. 26:720–721.
- Dijkstra KDB, Kalkman VJ, Dow RA, Stokvis FR, Van Tol J. 2014. Redefining the damselfly families: a comprehensive molecular phylogeny of Zygoptera (Odonata). Syst Entomol. 39:68–96.
- Dumont HJ, Vierstraete A, Vanfleteren JR. 2010. A molecular phylogeny of the Odonata (Insecta). Syst Entomol. 35:6–18.
- Feindt W, Fincke O, Hadrys H. 2014. Still a one species genus? Strong genetic diversification in the world's largest living odonate, the Neotropical damselfly Megaloprepus caerulatus. Conserv Genet. 15:469–481.
- Feindt W, Herzog R, Osigus HJ, Hadrys H. 2016. Short read sequencing assembly revealed the complete mitochondrial genome of Ischnura elegans (Vander Linden, 1820). Mitochondrial DNA part B (in press). DOI: 10.1080/23802359.2016.1192510
- Fincke OM, Hadrys H. 2001. Unpredictable offspring survivorship in the damselfly, Megaloprepus coerulatus, shapes parental behavior, constrains sexual selection, and challenges traditional fitness estimates. Evolution. 55:762–772.
- Fincke OM, Hedström I. 2008. Differences in forest use and colonization by Neotropical tree-hole damselflies (Odonata: Pseudostigmatidae): Implications for forest conversion. Stud Neotrop Fauna Environ. 43:35–45.
- Groeneveld LF, Clausnitzer V, Hadrys H. 2007. Convergent evolution of gigantism in damselflies of Africa and South America? Evidence from nuclear and mitochondrial sequence data. Mol Phylogenet Evol. 42:339–346.
- Hadrys H, Balick M, Schierwater B. 1992. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol. 1:55–63.
- Herzog R, Osigus HJ, Feindt W, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the emperor dragonfly Anax imperator LEACH, 1815 (Aeshnidae: Odonata) via NGS sequencing. Mitochondrial DNA part B (in press). DOI: 10.1080/23802359.2016.1186523
- Ingley SJ, Bybee SM, Tennessen KJ, Whiting MF, Branham MA. 2012. Life on the fly: phylogenetics and evolution of the helicopter damselflies (Odonata, Pseudostigmatidae). Zool Scripta. 41:637–650.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondrial DNA. 25:247–248.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Simon S, Hadrys H. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69:393–403.
- Swofford DL. 2002. PAUP* phylogenetic analysis using parsimony (*and other methods). Sunderland (MA): Sinauer Associates.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A. 2014. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166.
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2014. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA. 27:2271–2273.
