Abstract
Anthus hodgsoni is a species of small passerine bird in the family Motacillidae, which is widely distributed. In this study, we determined the complete mitochondrial genome of A. hodgsoni. The result showed that the total length of the mitochondrial genome was 16,886 bp and contained 2 ribosomal RNA genes, 22 transfer RNA genes, 13 protein-coding genes and 1 control region. The phylogenetic tree was reconstructed using the Bayesian analysis method and divided into four genera, Anthus, Dendronanthus, Motacilla and Tmetothylacus. The A. hodgsoni which we determined was clustered into genus Anthus and received strong support.
The olive-backed pipit (Anthus hodgsoni), classified as Least Concerned (LC) in the International Union for the Conservation (IUCN) Red List, is a small passerine bird of the pipit (Anthus) genus, which breeds across South, North Central and East Asia, as well as in the northeast of European Russia. They forage singly or in pairs on meadows and mainly feed on insects but also take on grass and weed seeds (MacKinnon et al. Citation2000). In the present study, the A. hodgsoni sample was collected from Huaining, Anqing of Anhui province, China (N 30°43′22.35″, E 116°49′27.68″). Now the specimen was deposited in the Laboratory of Evolution and Ecology, School of Life Sciences, Anhui University. Mitochondrial genome sequences have been proven to be useful for phylogenetic relationships at several taxonomic levels (Kumazawa & Endo Citation2004). We expect our result to provide a useful database for further research on A. hodgsoni.
The total genomic DNA of A. hodgsoni was extracted from muscle tissue using the standard proteinase-K/phenol–chloroform protocol (Sambrook & Russell Citation2006). Complete mitochondrial genome was amplified by polymerase chain reaction (PCR) using 16 pairs of primers then submitted it to GenBank (accession KX189345). The complete mitochondrial genome sequence of A. hodgsoni is 16, 886 bp in length and containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and 1 control region. The ND6 subunit gene and eight tRNA genes (tRNAPro, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer and tRNAGlu) were encoded on the L-strand, the remaining genes were encoded on the H-strand. The gene arrangement is similar to the complete mitochondrial genomes of other Fringillidae species (Lerner et al. Citation2011).
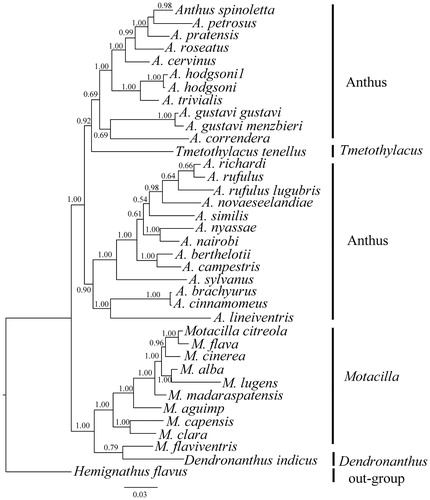
Phylogenetic relationship of 35 Motacillidae species were analyzed with the Bayesian inference (BI) method using the MrBayes version 3.1.2 software (Ronquist & Huelsenbeck Citation2003) based on nucleotide sequences of ND2 and cytb gene, selecting Hemignathus flavus as an outgroup. In this process, the best-fitting nucleotide substitution model (GTR + I + G) was selected via MrModeltest version 2.1 (Nylander Citation2004); the four independent Markov chain runs for 1,000,000 metropolis-coupled Monte Carlo (MCMC) generations, sampling every 1000 generations. When the average standard deviation of split frequencies reached a value less than 0.01, the first 1000 trees were discarded as “burn-in” and the remaining trees were used to calculate the Bayesian posterior probabilities (Pan et al. Citation2015). From the Bayesian analysis, the phylogenetic tree is divided into four genera and these relationships received strong support, with the topology similar to other research (). The first clade was genus Anthus, included 24 species, of which A. hodgsoni1 and A. hodgsoni were clustered into the same clade which indicate that the species of our study was A. hodgsoni. The A. hodgsoni appears to be very closely related to Anthus trivialis. However, Tmetothylacus tenellus was deeply nested within Anthus may infer that this species was belonged to Anthus. The second clade is genus Motacilla and genus Dendronanthus form the last clade, moreover they are the sister group to the genus Anthus and Tmetothylacus (Alström et al. Citation2015).
Figure 1. Inferred phylogenetic relationships among Motacillidae based on the ND2 and Cytb gene using Bayesian inference (BI). Numbers at each node indicate percentages of Bayesian posterior probabilities (BPPs). GenBank accession numbers for ND2 and Cytb are: Anthus spinoletta (None, LN650643), A. petrosus (None, APU46772), A. pratensis (AY259440, APU46774), A. roseatus (KJ455325, KJ456195), A. cervinus (None, ACU46776), A. hodgsoni1 (KJ455323, KJ456193), A. hodgsoni (KX189345), A. trivialis (KP671568, None), A. gustavi gustavi (HM538381, None), A. gustavi menzbieri (HM538396, None), A. correndera (KP671555, None), A. richardi (None, ARU46770), A. rufulus (None, KJ456194), A. rufulus lugubris (KJ455324, None), A. novaeseelandiae (KC545397, KC545397), A. similis (KJ455326, J456196), A. nyassae (None, JQ796089), A. Nairobi (None, AM231754), A. berthelotii (None, EU047723), A. campestris (JN614730, JN614900), A. Sylvanus (KJ455327, KJ456197), A. brachyurus (None, AF526450), A. cinnamomeus (AY329410, AY329447), A. lineiventris (KP671561, None), Motacilla citreola (KJ455509, AF526442), M. flava (KC759314, AF526468), M. cinerea (KJ455508, NC027933), M. alba (NC029229, EU167005), M. lugens (NC029703, NC029703), M. madaraspatensis (KJ455510, KJ456348), M. aguimp (GQ369693, AF526466), M. capensis (AY329427, AY329464), M. clara (KP671575,AF526469), M. flaviventris (None, AF526446), Tmetothylacus tenellus (KP671579, None), Dendronanthus indicus (KP671571, None) and Hemignathus flavus (KM078780, KM078780).
Funding information
This research was supported by the Foundation for College Student Innovation & Venture Project of Anhui University (201310357033).
Acknowledgements
The authors sincerely thank Xing Kang, Ke Fang, Chencheng Wang, Xiaonan Sun and Yanan Zhang for their assistance in this study.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
References
- Alström P, Jønsson KA, Fjeldså J, Ödeen A, Ericson PGP, Irestedt M. 2015. Dramatic niche shifts and morphological change in two insular bird species. R Soc Open Sci. 2:140–364.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Kumazawa Y, Endo H. 2004. Mitochondrial genome of the Komodo dragon: efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res. 11:115–125.
- Lerner HR, Meyer M, James HF, Hofreiter M, Fleischer RC. 2011. Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Curr Biol. 21:1838–1844.
- MacKinnon J, Phillipps K, He F-Q, Showler D, MacKinnon D. 2000. A field guide to the birds of China. Oxford (UK): Oxford University Press.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Pan T, Wang H, Hu C, Sun Z, Zhu X, Meng T, Meng X, Zhang B. 2015. Species delimitation in the Genus Moschus (Ruminantia: Moschidae) and its high-plateau origin. PLoS One. 10:e0134183.
- Sambrook J, Russell DW. 2006. Purification of nucleic acids by extraction with phenol: chloroform. CSH Protoc. 2006:pii:pdb.prot4455. http://dx.doi.org/10.1101/pdb.prot4455.
