Abstract
The complete mitochondrial genome of Perny's long-nosed squirrel (Dremomys pernyi) was firstly sequenced and characterized. The genome was 16,573 bp in length, and its composition and arrangement of genes were analogous to other rodents. The sequences of 13 protein-coding genes were used to construct phylogenetic tree for D. pernyi and other 13 sciurid species available on GenBank. To date, this is the first species whose complete mitochondrial genome sequence was sequenced in genus Dremomys. Our results will provide information for further molecular studies.
Genus Dremomys contains six species (Thorington & Hoffmann Citation2005), among them five species are distributed in China (Smith et al. Citation2008). The recent research demonstrates that Dremomys is not monophyletic, the species D. everetti is not clustered with other Dremomys species (Hawkins et al. Citation2016). To date, there are no complete mitochondrial genome data of species in this genus. In order to provide more molecular information for species in this genus, we first sequenced and characterized the complete mitochondrial genome of D. pernyi collected from Kangding of Sichuan province, China. The voucher specimen (G10138) was stored in School of Life Sciences, Guangzhou University.
According to our result, the mitogenome of D. pernyi was 16,573 bp in length and has been deposited in GenBank under the accession number KP708711. It contains 37 genes arranged in the same order with other reported vertebrate mitogenomes, including 22 tRNA genes, 13 protein-coding genes (PCGs), 2 rRNA genes and 2 non-coding regions. Among these genes, 28 are transcribed on the heavy-coding strand (H-strand), whereas the other nine genes on the light-coding strand (L-strand). The nucleotide composition is biased towards A and T with 59.1% A + T content (A = 31.9%, C = 27.9%, G = 13.0%, T = 27.2%). All PCGs are encoded on the H-strand except ND6. ATG is most preferred for most PCGs as the initial codon, whereas ND3 and ND5 start with ATA, and ND2 starts with ATT. ND6 and Cytb end with AGG and AGA, respectively, and other PCGs end with TAA or TAG, COX3 and ND4 end with TA- and T-, respectively, which can be completed by post-transcriptional polyadenylation (Ojala et al. Citation1981). The two rRNA genes (12S rRNA and 16S rRNA) are separated by gene tRNA-Val.
A set of 22 tRNA genes which arrange from 63 to 73 bp are found in the mitochondrial genome. All the tRNA genes could be folded into canonical cloverleaf structure except for tRNA-Ser(AGY) in which its dihydrouridine (DHU) simply forms a loop. The mitochondrial genome contains two non-coding regions, the 1098 bp control region and the 30 bp OL region. The control region was flanked by genes tRNA-Pro and tRNA-Phe, the OL region is located between tRNA-Asn and tRNA-Cys in a cluster of five tRNA genes (WANCY region).
The phylogenetic tree constructed by the Maximum Likelihood (ML) method using RAxML v7 (Stamatakis Citation2006) included mitochondrial genomes of13 PCGs of 13 species in family Sciuridae and two sequences of Gliridae downloaded from GenBank, and Ochotona curzoniae was used to root the phylogenetic tree. The result shows that D. pernyi is a sister lineage to genus Tamiops (). D. pernyi, T. maritimus, T. swinhoei and Callosciurus erythraeus form a well-supported lineage which belongs to tribe Callosciurini (Thorington & Hoffmann Citation2005).
Figure 1. Maximum likelihood (ML) phylogenetic tree based on complete mitochondrial genome under GTR + G model. ML bootstrap values are shown above nodes.
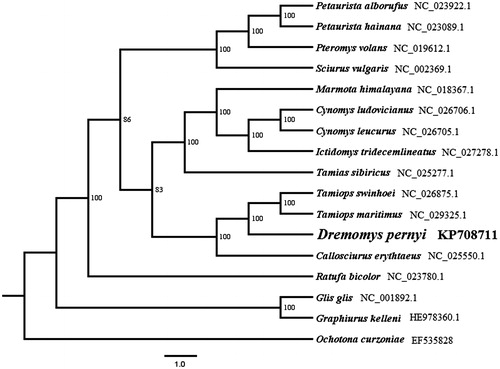
Molecular studies show good resolution on phylogenetic analysis and species identification in Dremomys (Hawkins et al. Citation2016; Li et al. Citation2008). The complete mitochondrial genome of D. pernyi reported in this study firstly provides the complete mitochondrial genome of this genus, it will be useful for molecular studies regarding this species in the future.
Funding information
The authors alone are responsible for the content and writing of the article. This research was funded by the Major International (Regional) Joint Research Project of NSFC (Grant No. 31110103910).
Acknowledgements
The authors thank the anonymous reviewers for providing valuable comments on the manuscript.
Disclosure statement
The authors report no conflicts of interest.
References
- Hawkins MT, Helgen KM, Maldonado JE, Rockwood LL, Tsuchiya MT, Leonard JA. 2016. Phylogeny, biogeography and systematic revision of plain long-nosed squirrels (genus Dremomys, Nannosciurinae). Mol Phylogenet Evol. 94:752–762.
- Li S, Yu F, Yang SH, Wang YX, Jiang XL, Mcguire PM, Feng Q, Yang JX. 2008. Molecular phylogeny of five species of Dremomys (Rodentia: Sciuridae), inferred from cytochrome b gene sequences. Zool Scripta. 37:349–354.
- Ojala DM, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Smith AT, Xie Y, Hoffmann RS, Lunde D, MacKinnon J, Wilson DE, Wozencraft WC. (2008). A guide to the mammals of China. Princeton (NJ): Princeton University Press. p. 51–54.
- Stamatakis A. 2006 . RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Thorington RW Jr, Hoffmann RS. (2005). Family Sciuridae. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Baltimore: The John Hopkins University. p. 754–818.
