Abstract
Hieroglyphus tonkinensis (Orthoptera: Caelifera: Acrididae) is an important agricultural pest to bamboo, rice and other gramineous crops. The complete mitochondrial genome of H. tonkinensis is 15,625 bp in length and consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and 1 A + T-rich region. The gene order of the mitogenome is identical with most orthopteran insects. Most protein-coding genes start with typical ATN codon except for COX1, which initiates with ACC codon instead. While all PCGs use complete stop codons (TAA and TAG). In addition, 13 related species and 2 outgroup taxa were used to construct the phylogenetic tree to further validate the mitogenome of H. tonkinensis. The result showed that H. tonkinensis is sister group to a clade of Oxya and Pseudoxya.
Hieroglyphus tonkinensis (Orthoptera: Caelifera: Acrididae) with gregariousness, flying force, move quickly, jumping strong, omnivorous and other characteristics, is a pest of bamboo, cane, rice, wheat, corn and other cereal crops. The insect is mainly distributed in Taiwan, Guangdong, Guizhou, Fujian, Hainan provinces and Guangxiautonomous regions in China (Lu et al. Citation2011). According to the Orthoptera Species File Online, Acrididae, as a subfamily, is included in the Acridoidea (Eades et al. Citation2015).
Mitochondrial DNA, as an effective molecular marker, is commonly used to investigate the population structure, phylogeography and phylogenetic analyses of insects (Ma et al. Citation2012; Zhang et al. Citation2013). However, the mitochondrial genome of H. tonkinensis is not available at present. Specimens of H. tonkinensis were collected from LiuwanDashan in Yulin City (Guangxi, China; N22°33′N, 109°46′E). the specimens were preserved in 100% absolute ethanol and stored in −20 °C freezer in Institute of Zoology of Shaanxi Normal University (accession No. M0683). Total genomic DNA was extracted from the muscle of one of the specimen’s femurs by the standard proteinase K and phenol/chloroform extraction method, then stored at −20 °C. Here, we report a complete mitochondrial genome of H. tonkinensis, which is 15 625 bp in length and has been deposited in GenBank (accession No. KX170936). It contains 13 typical protein-coding genes, 22 tRNA genes, 2 rRNA genes and 1 A + T-rich region, which is like the other orthopteran insects. The gene arrangement was identical with Calliptamus italicus (GenBank accession No.: NC_011305; Fenn et al. Citation2008). The overall base composition of the whole mitochondrial genome was A (42.9%), T (31.3%), C (15.1%) and G (10.8%), with an AT bias of 74.2%, as generally reported in other orthopteran mitogenomes. The 22 tRNA genes which were predicted by online software tRNAScan-SE (Lowe & Eddy Citation1997), rank from 62 bp (tRNACys) to 72 bp (tRNAVal) in length. Gene overlaps in the mitogenome of H. tonkinensis in a total of 41 bp in six locations with length in 1 ∼ 8 bp. The longest overlaps occur between trnW/trnC and trnY/COX1. The mitogenome has 21 intergenic spacers in a total of 146 bp with length varying from 1 to 35 bp.
All tRNAs except tRNASer(AGN) could be folded into typical cloverleaf secondary structures. Two rRNA genes were 1322 bp (lrRNA) and 795 bp (srRNA) long, which are located between tRNALeu(CUN) and A + T-rich region, separated by tRNAVal. Only COX1 uses ACC as a start codon, but the other protein-coding genes begin with the canonical ATN codon. While all the 13 PCGs use complete stop codons (TAG and TAA). The A + T-rich region, which is located between srRNA and tRNAIle, is 774 bp in length and has 85.3% AT content.
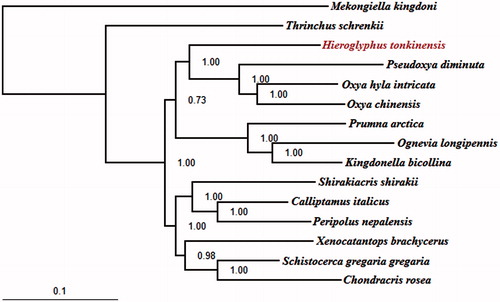
To further validate the mitogenome of H. tonkinensis, the phylogenetic analyses were performed using MrBayes 3.1.2 (Ronquist & Huelsenbeck Citation2003) based on the concatenated dataset (PCGs) of mitogenomes of H. tonkinensis and the other 15 taxa that were retrieved from GenBank, including 13 Acrididae ingroup and two outgroup taxa ().The result showed that H. tonkinensis is sister group to a clade of Oxya and Pseudoxya.
Figure 1. The BI phylogenetic tree of the Hieroglyphus tonkinensis in this study and other 13 related species and 2 outgroup taxa based on mitochondrial PCGs concatenated dataset. Note: Mekongiella kingdoni: NC_023921; Thrinchus schrenkii: NC_014610. The 2 outgroup taxa belong to Acridoidea. Hieroglyphus tonkinensis: KX170936; Pseudoxya diminuta: NC_025765; Oxya hyla intricata: KP313875; Oxya chinensis: NC_010219; Prumna arctica: NC_013835; Ognevia longipennis: NC_013701; Kingdonella bicollina: NC_023920; Shirakiacris shirakii: NC_021610; Calliptamus italicus: NC_011305; Peripolus nepalensis: NC_029135; Xenocatantops brachycerus: NC_021609; Schistocerca gregaria gregaria: NC_013240; Chondracris rosea: NC_019993. The 13 related species all belong to Acrididae.
Disclosure statement
The authors alone are responsible for the content and writing of the article, and has no competing interests.
Funding information
This project was supported by National Natural Science Foundation of China (No. 31372192).
References
- Eades DC, Otte D, Cigliano MM, Braun H. 2015. Orthoptera species file. Version 5.0/5.0. [Internet] [cited 2015 May 3]. Available from: http://Orthoptera.SpeciesFile.org
- Fenn JD, Song H, Cameron SL, Whiting MF. 2008. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data[J]. Mol Phylogenet E. 49:59–68.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Lu F, Zhao D, Huang Liguang, Wang A, Chen Q. 2011. Biological characteristics and effective control of Hieroglyphus tonkinensis I.Bol in Hainan[J]. Chinese J. Tropical Crops. 32:1144–1149.
- Ma C, Yang P, Jiang F, Chapuis MP, Shali Y, Sword GA, Kang L. 2012 . Mitochondrial genomes reveal the global phylogeography and dispersal routes of the migratory locust. Mol Ecol. 21:4344–4358.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Zhang HL, Zeng HH, Huang Y, Zheng ZM. 2013. The complete mitochondrial genomes of three grasshoptters, Asiotmethis zacharjini, Filchnerella helanshanensis and Pseudotmethis rubimarginis (Orthoptera: Pamphagidae). Gene. 517:89–98.
