Abstract
In this study, we first present the complete mitochondrial genome of Hemipristis elongata, the only member of genus Hemipristis in family Hemigaleidae. It is 16,691 bp in length with the typical gene order invertebrates. Its overall base composition is 31.7% A, 24.1% C, 12.9% G and 31.3% T. Two start codons (ATG and GTG) and three stop codons (TAG, AGA and TAA/T) are found in the protein-coding genes. The 22 tRNA genes ranged from 67 bp (tRNA-Cys, tRNA-Ser2) to 75 bp (tRNA-Leu1). The phylogenetic result showed that H. elongata was clustered to Hemigaleus microstoma and formed Hemigaleidae.
The snaggletooth shark (Hemipristis elongata) is the only member of genus Hemipristis in family Hemigaleidae. It has a slender body, commonly in habiting in inshore and offshore on the continental and insular shelves of Indo-West Pacific (Compagno Citation1984; Carpenter et al. Citation1997). The reproductive modes of H. elongata is viviparous, with 2–11 young in a litter after a gestation period of 7–8 months (Last & Stevens Citation1994). In this study, we determined the complete mitochondrial genome of H. elongata for the first time, and constructed the phylogenetic tree using the mitogenomes in Carcharhiniformes.
One specimen of H. elongata was captured in Ranong, Thailand and preserved in the Museum of Marine Biology in Wenzhou Medical University with voucher: RN2012122416. The experimental protocol and data analysis methods followed Chen et al. (Citation2014). Including H. elongata, twenty-nine species of Carcharhiniformes with the complete mitogenomes available in the GenBank were selected to construct the phylogenetic tree. The Bayesian method was fulfilled with the GTR + I + G model by three partitions of the mitogenomic data:12S and 16S rRNA genes, and the first and second codons of the 12 protein-coding genes (except ND6 gene).
The complete mitochondrial genome of H. elongata is 16,691 bp in length (Genbank Accession Number: KU508621). It contains 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes and a control region. The gene composition, arrangement and transcriptional orientation are identical to most mitogenomes of vertebrates. Its nucleotide base composition is 31.7% A, 24.1% C, 12.9% G and 31.3% T. There were total 23 bp short intergenic spaces located in 12 gene junctions and 33bp overlaps located in 10 gene junctions in the mitogenome. Both 12S rRNA (953 bp) and 16S rRNA (1666 bp) genes were between tRNA-Phe and tRNA-Leu1 genes, separated by tRNA-Val gene. All protein-coding genes started with the standard ATG codon except CO1 gene, which used GTG as the start codon. Except for the ND6 gene using AGA as the stop codon, the remaining protein-coding genes were terminated by the typical TAG or TAA/T codon. Twenty-two tRNA genes interspersed between the rRNA and protein-coding genes, ranging from 67bp (tRNA-Cys and tRNA-Ser2) to 75 bp (tRNA-Leu1). Except for the tRNA-Ser2 that replaced the dihydrouridine arm by a simple loop, the remaining tRNAs could fold into a typical clover-leaf secondary structure. The control region was 1063 bp in length, with the rich in A + T (67.1%) and poor in G (12.6%) content.
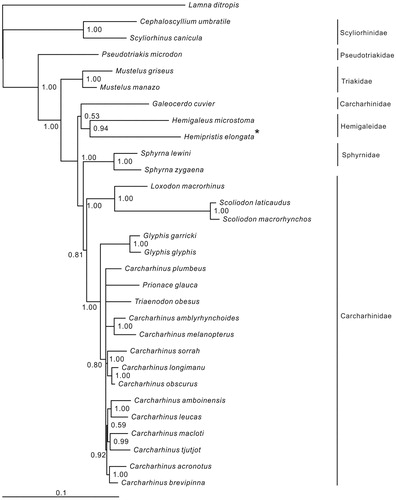
Six families of Carcharhiniformes were included in the phylogenetic tree. Most nodes of the Bayesian tree were well supported (). The relationship of three basal families (Scyliorhinidae, Pseudotriakidae and Triakidae) was clear. Hemipristis elongata was clustered to Hemigaleus microstoma with high support value (94%), however, this clade (Hemigaleidae) was then clustered to Galeocerdo cuvier (Carcharhinidae) with low support value (53%). In addition, Sphyrnidae were inserted between two main clades of (G. cuvier + Hemigaleidae) and the remaining Carcharhinidae species, it demonstrated that Carcharhinidae is paraphyletic. Therefore, the relationship of the families (Carcharhinidae, Hemigalidae and Sphyrnidae) in Carcharhiniformes need further studies.
Figure 1. Phylogenetic position of Hemipristis elongata. Lamna ditropis (KF962053.1) was selected as the out group. The twenty-nine species from the order Carcharhiniformes were: Carcharhinus acronotus (NC_024055.1), C. amblyrhynchoides (NC_023948.1), C. amboinensis (NC_026696.1), C. brevipinna (KM244770.1), C. leucas (KF646785.1), C. longimanu (NC_025520.1), C. macloti (NC_024862.1), C. melanopterus (NC_024284.1), C. obscurus (NC_020611.1), C. plumbeus (NC_024596.1), C. sorrah (NC_023521.1), C. tjutjot (KP091436.1) Galeocerdo cuvier (NC_022193.1), Loxodon macrorhinus (KT347599), Prionace glauca (NC_022819.1), Scoliodon laticaudus (KP336547.1), S. macrorhynchos (NC_018052.1), Triaenodon obesus (KJ748376.1), Glyphis glyphis (NC_021768.2),G. garricki (KF646786.1), Mustelus griseus (NC_023527.1), M. manazo (NC_000890.1), Cephaloscyllium umbratile (KT003686), Hemigaleus microstoma (KT003687), Hemipristis elongata (KU508621), Scyliorhinus canicula (NC_001950.1), Pseudotriakis microdon (NC_022735.1), Sphyrna lewini (NC_022679.1), Sphyrna zygaena (NC_025778.1).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding information
This study was supported by the Special Scientific Research Funds for Central Non-profit Institutes, Chinese Academy of Fishery Sciences (2015B02XK02) and Guangdong Natural Science Foundation (2015A030310002).
References
- Carpenter KE, Krupp F, Jones DA, Zajonz U. 1997. FAO species identification field guide for fishery purposes. In: Living marine resources of Kuwait, Eastern Saudi Arabia, Bahrain, Qatar, and the United Arab Emirates. Rome: FAO. p. 293.
- Chen X, Ai W, Xiang D, Chen S. 2014. Complete mitochondrial genome of the red stingray Dasyatis akajei (Myliobatiformes: Dasyatidae). Mitochondrial DNA. 25:37–38.
- Compagno LJV. 1984. FAO fisheries synopsis no. 125. In: FAO species catalogue. vol. 4. Sharks of the world. An annotated and illustrated catalogue of shark species known to date. Part 2: Carcharhiniformes. Rome: FAO. p. 251–655.
- Last PR, Stevens JD. 1994. Sharks and rays of Australia. Australia: CSIRO. p. 513.
