Abstract
Conyza blinii is a widely used medicinal plant in the southwest of Sichuan with great economic value. Its chloroplast (cp) genome was 152,503 bp in length with the typical quadripartite structures of the large (LSC, 84,210 bp) and small (SSC, 18,257 bp) single-copy regions, separated by a pair of inverted repeats (IRs, 25,018 bp). The genome contains 114 encoding genes, including 80 protein-coding genes, 30 tRNAs and 4 rRNAs. The overall GC content of the whole cp genome is 37.4% which is similar to the other reported asteridae cp genomes.
Chloroplasts (cps) are indispensable plant cell organelles that conduct photosynthesis in the presence of daylight. The chloroplast genome in land plants is a circular molecule ranging in size from 76 to 217 kb with a highly conserved structure of two copies of large inverted repeats (IR) detached by small (SSC) and large (LSC) single-copy regions. Characteristic of the cp genome have been used comprehensively as a tool to investigate evolutionary relationships and molecular phylogenetic of seed plants (Saski et al. Citation2005; Lee et al. Citation2006).
Conyza blinii is a biennial herbaceous plant, belongs to Asteraceae which is a larger family in angiosperms, and has been used in folk medicine for a long time in the south-west of China. With the further study, a new neoclerodane diterpenelactone, blinin, was isolated from the whole plant of C. blinii. It is also reported to have a decisive role in pharmaceutical research (Yang et al. Citation1989).
This study reports the sequencing of the C. blinii, and the plants in the wild were collected in Panzhihua (Coordinate: 26.562222N, 101.796389E; Altitude: 1910 m). The whole cp genome was sequenced using HiSeq 2000, Illumina and assembled into the complete cp genome using Sequencher version 4.10. DualOrganellarGenoMe Annotator (Austin, TX) (Wyman et al. Citation2004) was utilized to annotate the cp genome with BLASTX and BLASTN identifying the location of encoding genes and RNAs. The complete genome sequence was submitted to GenBank under the accession number KX085421.
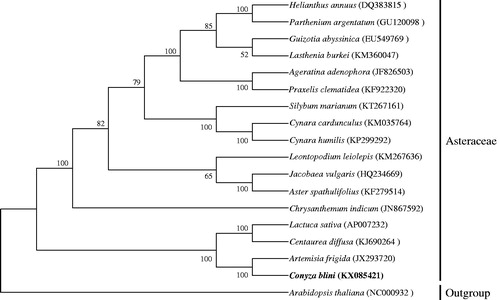
The total size of complete cp genome was 152,503 bp with 37.4% GC content, consisted of a typical quadripartite structures of one LSC (84,210 bp), one SSC (18,257 bp), and two IRs (25,018 bp). The GC content of the SSC (43%) was higher than that of LSC and IR (35.3% and 31.4%, correspondingly), which has significant differences compared to the majority of angiosperm (Liu et al. Citation2013; Zhang et al. Citation2014; Choi & Park Citation2015; Curci et al. Citation2015; Lu et al. Citation2015). Usually, the IR region has the highest GC content, mainly caused by the the high GC content of the four ribosomal RNA (rRNA) genes. A total of 114 genes were successfully annotated, including 80 protein-coding genes, 30 tRNA genes and 4 rRNA genes; and 21 out of 114 genes were duplicated in inverted repeat regions. There were 17 intron-containing genes, almost all of which were single-intron genes except for ycf3 and clpP with two introns separately. Exceptionally, rps12 was a reverse splicing gene, of which the 5′ end exon located in LSC region and 3′ end exon was in IR region. The phylogenetic relationship of C. blinii was deduced by comparing it with the other 18 chloroplast genomes downloaded from Genbank ().
Funding information
This work was supported by funding from the Department of Science and Technology of Sichuan Province (contract number:GR-2013-C-04).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Choi KS, Park S. 2015. The complete chloroplast genome sequence of Aster spathulifolius (Asteraceae); genomic features and relationship with Asteraceae. Gene. 572:214–221.
- Curci PL, De Paola D, Danzi D, Vendramin GG, Sonnante G. 2015. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. PLoS One. 10:e0120589.
- Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. 2006. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 7:61.
- Liu Y, Huo N, Dong L, Wang Y, Zhang S, Young HA, Feng X, Gu YQ. 2013. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One. 8:e57533.
- Lu C, Shen Q, Yang J, Wang B, Song C. 2015. The complete chloroplast genome sequence of Safflower (Carthamus tinctorius L.). Mitochondrial DNA. 27:3351–3353.
- Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. 2005. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol. 59:309–322.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang CR, He ZT, Li XC, Zheng QT, He CH, Yang J, Morita T. 1989. Blinin, a neoclerodane diterpene from Conyza blinii. Phytochemistry. 28:3131–3134.
- Zhang Y, Li L, Yan TL, Liu Q. 2014. Complete chloroplast genome sequences of Praxelis (Eupatorium catarium Veldkamp), an important invasive species. Gene. 549:58–69.