Abstract
The complete mitochondrial genome of the Critically Endangered Trinidad Piping Guan, Pipile pipile (Jacquin 1784) synonym Aburria pipile was sequenced for the first time in this study. The genome is 16,665 bp in length with overall base compositions of 30.1, 23.7, 32.3 and 13.9% for A, T, C, and G, respectively. Structurally, the P. pipile mitogenome is comparable to that of other Galliformes, thereby demonstrating typical avian gene organization. The mitogenome was subsequently used to produce a revised phylogenetic placement of P. pipile within the Galliforme order, positioning the Pipile genus basal within the Cracidae family. It is further envisaged that this novel genomic data will contribute to a wider understanding of genetic relationships within the genus Pipile and the analysis of the evolutionary relationships of the Galliforme order in a wider avian context.
Mitogenome announcement
The Trinidad Piping Guan Pipile pipile is a Critically Endangered Cracid (Galliforme) endemic to the island of Trinidad, with an estimated population of less than 200 individuals (Hayes et al. Citation2009a). The population has been in ongoing decline largely due to anthropogenic pressures (Hayes et al. Citation2009a, Citation2009b). The IUCN and BirdLife International recognize Trinidad Piping Guan as a species of concern, classified as Critically Endangered, protected under Appendix I of the Convention on the International Trade in Endangered Species (BirdLife International Citation2015; CITES Citation2015; IUCN Citation2015). This mitochondrial genome represents the novel use of genetic data sourced from contemporary Trinidad Piping Guans from the wild on the island of Trinidad.
Samples were sourced from wild Trinidad Piping Guans (P. pipile) in the Matura Forest Reserve, Northern Range, Trinidad. P. pipile mitochondrial genomes were amplified from the feathers of live birds and liver tissue from a nonviable embryo.
The complete mitochondrial genome of P. pipile, sequenced for both heavy and light strands, was 16,665 bp ±2 bp in length (Genbank: KU221051, KU221052, KU221053), 13 protein-coding genes, 2 rRNAs, and 22 tRNAs were identified demonstrating organization consistent with Galliforme mito-genomes. Base composition of the three complete genomes demonstrate an A + T bias (53.8%), where overall base composition of the genome were as follows; A (30.1%), T (23.7%), C (32.3%), and G (13.9%).
All compositional features of the genome were consistent between samples, with length variation occurring due to indels in the hypervariable sections of the control region. The majority of genes are encoded on the heavy strand, however, the ND6 gene and eight tRNA genes are light strand encoded, as in other Galliformes. The majority of genes are contiguous within the sequence, however, gene overlaps occur in four locations; tRNAGln and tRNAMet, tRNACys and tRNATyr, ATP8 and ATP6, and ND4L and ND4. Additionally, noncoding intergenic spacers of variable length (1–13nts) occur between some genes. The additional nontranslated nucleotide at position 174 of the ND3 gene was observed consistent with the observation of this additional nucleotide in other Galliformes (Mindell et al. Citation1998).
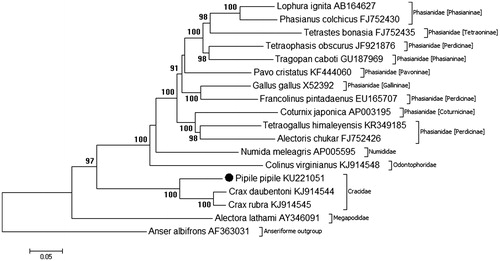
Phylogenetic analysis of the mitochondrial genome of P. pipile and other Galliforme species is shown in , constructed using MEGA7.0 (http://www.megasoftware.net/) (Kumar et al. Citation2016). The Pipile genus resolves basally within the Cracidae order of the Galliforme phylogenetic tree, with a high degree of bootstrap support (100%). Position of the Cracidae family within the Galliforme tree is consistent with initial mitogenome analysis (Crax sp.) in previous research (Meiklejohn et al. Citation2014). Basal divergence of the Pipile genus in relation to the Crax is consistent with individual mitochondrial gene analysis illustrated by Crowe et al. (Citation2006), however, full resolution of the Cracidae family is restricted by a lack of complete mitochondrial genome information for Cracid species. The complete mitogenome of the Trinidad Piping Guan, Pipile pipile, is used for the first time in phylogenetic analysis, providing essential molecular data and evolutionary information for further analysis of both the Pipile genus and the Cracidae family.
Figure 1. Phylogenetic relationships of Pipile pipile within the Galliforme order, inferred from whole mitochondrial genomes. Phylogenetic tree constructed using complete mitogenomes from Genbank in MEGA 7.0 using the Maximum-Likelihood method with 1000 bootstrap replicates. Genbank accession numbers are given after the species name, bootstrap support is given at each node.
Acknowledgements
The authors would like to acknowledge Prof. J. Cooper and the Pawi Study Group for their assistance in acquisition of verified reference samples from wild Trinidad Piping Guans.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
References
- Birdlife International. 2015. BirdLife International Species Information Pipile pipile [Internet]. [cited 2015 Oct 10]. Available from: http://www.birdlife.org/datazone/speciesfactsheet.php?id =87.
- CITES. 2015. CITES Species Database [Internet]. [cited 2015 Oct 10]. Available from: http://speciesplus.net/#/taxon_concepts/7109/legal.
- Crowe TM, Bowie RCK, Bloomer P, Mandiwana TG, Hedderson TAJ, Randi E, Pereira SL, Wakeling J. 2006. Phylogenetics, biogeography and classification of, and character evolution in, gamebirds (Aves: Galliformes): effects of character exclusion, data partitioning and missing data. Cladistics. 22:495–532.
- Hayes F, Sanasie B, Samad I. 2009a. Status and conservation of the critically endangered Trinidad Piping-Guan Aburria pipile. Endanger Species Res. 7:77–84. [Internet]. [cited 2012 May 16]. Available from: http://www.int-res.com/abstracts/esr/v7/n1/p77-84/.
- Hayes F, Shameerudeen C, Sanasie B, Hayes B, Ramjohn C, Lucas F. 2009b. Ecology and behaviour of the critically endangered Trinidad Piping-Guan Aburria pipile. Endanger Species Res. 6:223–229.
- IUCN. 2015. The IUCN Red List of Threatened Species. Red List [Internet]. [cited 2015 Oct 10]. Available from: http://www.iucnredlist.org/details/22678401/0.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874.
- Meiklejohn KA, Danielson MJ, Faircloth BC, Glenn TC, Braun EL, Kimball RT. 2014. Incongruence among different mitochondrial regions: a case study using complete mitogenomes. Mol Phylogenet Evol. 78:314–323.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15:1568–1571.
