Abstract
The complete plastid genome (plastome) of Pentactina rupicola Nakai, the sole member of genus Pentactina, endemic to Korea, was determined in this study. The plastome of P. rupicola is 156,612 bp in length and is composed of a pair of 26,351 bp inverted repeat regions (IRa and IRb) separating large (LSC) and small (SSC) single-copy regions of 84,970 and 18,940 bp, respectively (NCBI acc. no. NC 016921). The plastome encodes 129 genes, of which 112 are unique, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. Seventeen genes contain one intron and the ycf3 gene has two introns. The second intron of clpP is absent in the P. rupicola plastome. The AT content of P. rupicola is 63% overall, and in the LSC, SSC, and IR regions is 65%, 69%, and 58%, respectively. A total of 63 simple sequence repeats (SSR) are distributed among the noncoding regions of the genome. Phylogenetic analysis of the combined 82-gene data set for 35 plastomes suggests that P. rupicola is sister to the Pyrusmalus clade.
Keywords:
Pentactina Nakai is a monotypic genus comprising P. rupicola Nakai, which is endemic to Korea (Nakai Citation1917). Pentactina rupicola is a perennial dwarf shrub that has a restricted distribution range in the Mt. Geumgang region of central Korea. It belongs to the family Rosaceae, which consists of approximately 90 genera and 3000 species (Potter et al. Citation2007). The genus Pentactina is classified as a member of the tribe Spiraeeae on the basis of its general morphology (Takhtadzhian Citation1997). The close relationship between Spiraea and Pentactina was suggested by a pollen morphology study (Lee et al. Citation1993). Lee and Hong (Citation2011) reported that Pentactina possibly has a close relationship with Petrophyton based on molecular data. Here, we present the complete plastome of P. rupicola.
Approximately 2 g of P. rupicola leaves were collected from the cultivated individuals. A voucher specimen was deposited in the Korea University Herbarium (KUS 2007-0740). Genomic DNA was isolated using the CTAB method (Doyle & Doyle Citation1987). The DNA was purified by equilibrium ultracentrifugation using CsCl-ethidium bromide gradients, and further purified using a dialysis membrane (Palmer Citation1986). A sample of the purified DNA was deposited in the Plant DNA Bank of Korea (PDBK 2007-0740). A combination of short- and long-range PCRs and continuous sequencing of PCR products using primer walking was utilized for whole plastome sequencing. The amplification primers were designed on the basis of the plastomes of Nicotiana, Panax, and Morus (Kim & Lee Citation2004; Yukawa et al. Citation2005; Ravi et al. Citation2006). The PCR products were purified using the MEGAquick-spin kit (iNtRON, Seoul, Korea), and the cleaned products were sequenced using a series of primers oriented in both directions at intervals of 300–800 bp, using an ABI 3730XL automatic sequencer. Sequence fragments were edited and assembled using Sequencher 4.7 (Gene Code Corporation, Ann Arbor, MI). Gene annotations were performed using BLAST of the National Center for Biotechnology Information (NCBI), DOGMA (Wyman et al. Citation2004), and tRNAscan-SE (Lowe & Eddy Citation1997).
The structure, gene content, gene order, and AT content of the P. rupicola plastome are similar to those of a typical angiosperm plastome (Kim & Lee Citation2004; Kim et al. Citation2009). The P. rupicola plastome is 156,612 bp in length, including a large single-copy (LSC) region of 84,970 bp and a small single-copy (SSC) region of 18,940 bp separated by two inverted repeat (IR) regions, each of 26,351 bp (NCBI acc. no. NC016921). The plastome contains 129 genes (112 unique), including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Six protein-coding genes, seven tRNA genes and four rRNA genes are duplicated in the IR regions. A total of 18 genes contain one (17 genes) or two (ycf3 gene) introns. One of the notable features of the P. rupicola plastome is the absence of the intron II of the clpP gene. Of the published Rosaceae plastomes (Terakami et al. Citation2012), the second intron of the clpP gene has been lost only in the P. rupicola plastome (Jansen et al. Citation2007).
The P. rupicola plastome comprises 57% coding regions (52% protein-coding and 5% RNA-coding regions) and 43% noncoding regions (11% intron and 32% intergenic spacers). The AT content in the noncoding regions (52%) is higher than that in the coding (48%) regions. The overall AT content of the plastome is 63%, and the AT contents of the LSC, SSC, and IR regions are 65%, 69%, and 58%, respectively. A total of 63 simple sequence repeat (SSR) loci, which can be defined as having more than 10 duplications of simple nucleotide(s), are distributed among the noncoding regions of the genome. Among these, the majority of the SSR loci (50/63) are mono-SSR. In addition, eight di-SSR loci, three tri-SSR loci, one tetra-SSR locus, and one penta-SSR locus were identified. Some of these loci will be useful in identifying cultivars of P. rupicola.
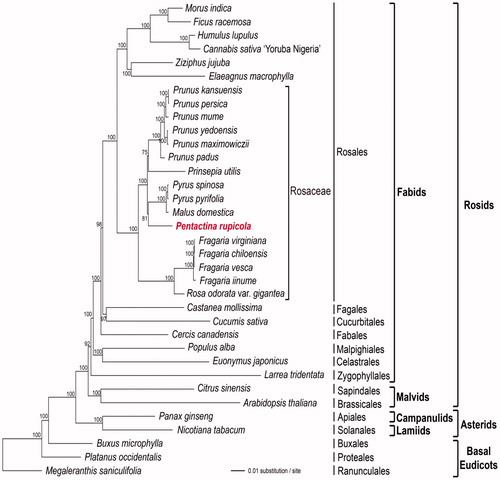
The phylogenetic relationships of P. rupicola, based on the data of 82 protein-coding and rRNA genes (aligned length: 77,096 bp) from 35 plastomes, were examined by performing maximum-likelihood (ML) analyses using RAxML v. 8.2.8 (Stamatakis Citation2014) with 1000 bootstrap replicates. The phylogenetic tree indicated that P. rupicola is most closely related to the Pyrusmalus clade in Rosaceae with bootstrap values of 81% ().
Figure 1. ML tree based on 82 protein-coding and rRNA genes from 35 plastid genomes as determined by RAxML (−ln L = −441,234.528222). The numbers at each node indicate the ML bootstrap values. Genbank accession numbers of used taxa are shown below, Arabidopsis thaliana (NC_000932), Buxus microphylla (NC_009599), Canabis sativa ‘Yoruba Nigeria’ (NC_027223), Castanea mollissima (NC_014674), Cercis canadensis (KF856619), Citrus sinensis (NC_008334), Cucumis sativus (NC_007144), Elaeagnus macrophylla (NC_028066), Euonymus japonicus (NC_028067), Ficus racemosa (NC_028185), Fragaria chiloensis (NC_019601), F. iinumae (NC_024258), F. vesca (NC_015206), F. virginiana (NC_019602), Humulus lupulus (NC_028032), Larrea tridentata (NC_028023), Malus domestica (Genome Database for Rosaceae), Megaleranthis saniculifolia (NC_012615), M. indica (NC_008359), N. tabacum (NC_001879), P. ginseng (NC_006290), P. rupicola (NC_016921, in this study), Platanus occidentalis (NC_008335), Populus alba (NC_008235), Prinsepia utilis (NC_021455), Prunus kansuensis (NC_023956), P. maximowiczii (NC_026981), P. mume (NC_023798), P. padus (NC_026982), P. persica (NC_014697), P. yedoensis (NC_026980), P. pyrifolia (NC_015996), P. spinosa (NC_023130), Rosa odorata var.gigantea (KF753637), and Ziziphus jujuba (NC_030299).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104:19369–19374.
- Kim K-J, Lee H-L. 2004. Complete chloroplast genome sequences from Korean Ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11:247–261.
- Kim Y-K, Park C-W, Kim K-J. 2009. Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia, and its evolutionary implications. Mol Cells. 27:365–381.
- Lee C, Hong S-P. 2011. Phylogenetic relationships of the rare Korean monotypic endemic genus Pentactina Nakai in the tribe Spiraeeae (Rosaceae) based on molecular data. Plant Syst Evol. 294:159–166.
- Lee S-T, Jeong Y-J, Lee J-H. 1993. Palynological relationship between Pentactina rupicola Nakai and its relative taxa. Korean J Plant Taxon. 23:149–159.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Nakai T. 1917. Notulæ ad plantas japoniæ et Coreæ. XIII. Bot Mag. 31:3–30.
- Palmer JD. 1986. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 118:167–186.
- Potter D, Eriksson T, Evans RC, Oh S, Smedmark JEE, Morgan DR, Kerr M, Robertson KR, Arsenault M, Dickinson TA, et al. 2007. Phylogeny and classification of Rosaceae. Plant Syst Evol. 266:5–43.
- Ravi V, Khurana JP, Tyagi AK, Khurana P. 2006. The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis. Tree Genet Genomes. 3:49–59.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Takhtadzhian AL. 1997. Diversity and classification of flowering plants. New York: Columbia University Press.
- Terakami S, Matsumura Y, Kurita K, Kanamori H, Katayose Y, Yamamoto T, Katayama H. 2012. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): genome structure and comparative analysis. Tree Genet Genomes. 8:841–854.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yukawa M, Tsudzuki T, Sugiura M. 2005. The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum). Plant Mol Biol Rep. 23:359–365.
