Abstract
The giant flying squirrel Petaurista petauri is a large rodent studied by few researchers. Here, we sequenced the complete mitochondrial genome of P. petauri. Similar to the typical vertebrate mitochondrial genome, the mtDNA of P. petauri also contained 37 genes (13 protein-coding genes, 2 rRNA genes, and 22 tRNA genes) and a noncoding region (D-loop). We also analyzed the phylogenetic relationship of P. Petauri to 14 other closely related species using the Bayesian inference. This work will contribute to our understanding of this species’ evolution and conservation.
The giant flying squirrel Petaurista petauri (also known as Petaurista yunnanensis) is a large rodent distributed mainly in the southwest of China, and likely in Burma and Laos as well (Smith & Xie Citation2008). Petaurista petauri (family Sciuridae) is a solitary, nocturnal mammal that prefers to inhabit caves, rock tunnels, and other holes (Smith & Xie Citation2008). As no research has been done on the molecular evolution of this species, we know little of their phylogenetic history, and the taxonomy of this species remains controversial (Smith & Xie Citation2008). Currently, only three complete sequences of the mitochondrial DNA of genus Petaurista have been published (Kong et al. Citation2015). Given their uncertain taxonomy and remarkable subterranean lifestyle, sequencing the mitochondrial genome sequence of P. petauri was a priority. We hope this work will contribute to resolving the taxonomic dispute around P. petauri and provide more valuable information about their life history.
One adult P. petauri was collected from the wild in Yunnan, China (coordinates: E 99°18.639′, N 27°35.702′). This specimen is stored at a museum in the College of Life Sciences, China West Normal University, China, with accession No. PP-15-01. We used standard phenol–chloroform methods to extract the genomic DNA from the muscle tissue (Sambrook & Russell Citation2001). After obtaining the complete mitochondrial genome of P. petauri, sequences were uploaded to the GenBank database with accession number KX528208. The complete mtDNA of P. petauri is 16620bp in length. It contains 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes for a total of 37 encoded genes, as well as a noncoding D-loop region. All genes were distributed on the H-strand except for the ND6 subunit gene and nine tRNA genes, which were encoded on the L-strand. Overall, the nucleotide base composition of the mtDNA is A (30.3%), G (14.7%), C (31.0%), and T (24.0%), indicating a strong AT bias (Shadel & Clayton Citation1997).
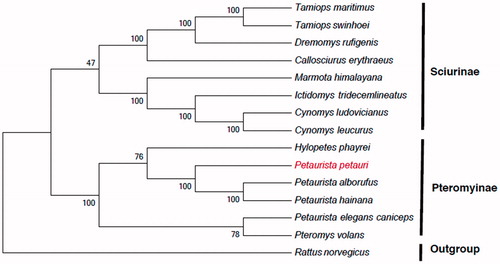
In order to resolve the taxonomic dispute surrounding P. petauri and understand its relationship to other species, we compared its mtDNA to that of 14 other species from the family Sciuridae to reconstruct the phylogenetic tree. Using jModelTest version 2.1.4 (Darriba et al. Citation2012), we evaluated several models for phylogenetic analysis of the nucleotide sequences. The best-fitting model was the General Time Reversible model (GTR) (Tavaré Citation1986) coupled with Gamma distributed with Invariant sites (G + I). The nucleotide sequences were used to generate a maximum-likelihood tree with bootstrapping (1000 replicates) using MEGA version 5 (Tamura et al. Citation2011). The phylogenetic tree is classified into two large clades and the outgroup (). The first lineage, containing six genera (Tamiops, Dremomys, Callosciurus, Marmota, Ictidomys, and Cynomys), belongs to the subfamily Sciurinae. The second lineage, subfamily Pteromyinae, includes the genera Hylopetes, Pteromys, and Petaurista. The outgroup was Rattus norvegicus. We expect that this work can contribute to the molecular identification of P. petauri and be helpful to explore the phylogeny of the Sciuridae.
Figure 1. Maximum-likelihood tree inferred from the nucleotide sequence of whole mitogenome. The numbers above branches specify bootstrap percentages (1000 replicates). GenBank accession numbers for the published sequences are H. phayrei (NC026443); P. volans (JQ230001); P. alborufus (NC023922); P. hainana (JX572159); P. elegans caniceps (KU579289); C. erythraeus (NC025550); C. leucurus (NC026705); C. ludovicianuse (NC026706); D. rufigenis (NC026442); I. tridecemlineatus (NC027278); M. himalayana (NC018367); T. maritimus (NC029325); T. swinhoei (NC026875); R. norvegicus (KF011917).
Acknowledgements
We are grateful to our field assistants Shuikai Feng. We thank Baimaxueshan Nature Reserve for our work permit.
Disclosure statement
The authors have declared no conflicting interests. The authors alone are responsible for doing the research and writing the paper.
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 9:772.
- Kong L, Wang Y, Cong H, Wang W, Li Y. 2015. Characterization of the complete mitochondrial genome of the Hainan giant flying squirrel Petaurista hainana (Rodentia: Sciuridae). Mitochondrial DNA. 26:871–872.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. New York: Cold Spring Harbour Laboratory Press.
- Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 66:409–435.
- Smith AT, Xie Y. (2008). A guide to the mammals of China, Xiii. In: Princeton NJ, editor. Princeton: Princeton University press.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Tavaré S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 17:57–86.
