Abstract
The complete mitogenome sequence of Batrachuperus pinchonii (Caudata: Hynobiidae) has been amplified and sequenced in this study. The overall base composition of B. pinchonii mitogenome is 33.9% for A, 19.7% for C, 13.6% for G, and 32.8% for T and has low GC content of 33.3%. The assembled mitogenome, consisting of 16,381 bp, has 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and 1 D-loop region. All PCGs, except for ND6 gene, were encoded on H-strand. Phylogenetic analysis with the whole mitogenome sequences revealed a close relationship of B. pinchonii with B. tibetanus.
Batrachuperus pinchonii (David), which belongs to Batrachuperus of Hynobiidae of Caudata, is an amphibian with tail adapted to the cold water of the alpine environment (Qi et al. Citation2010). This species is also an economic animal due to the medicinal value of enhancing fracture healing. Historical medicinal books of China had recorded B. pinchonii as Chinese medicine, and named Qianghuo Yu. This species has the effect of reunion of fractured tendons and bones, nourishing and strengthening the life, and has been used for the treatment of traumatic injury and joint pain (Du et al. Citation2015). Despite its useful applications, there is little report on molecular and genomic resources (Che et al. Citation2012; Chen et al. Citation2015). In this study, we characterized the mitogenome sequence of B. pinchonii.
The sample was collected from Yajiageng mountain (29°50′N, 102°20′E) in Kangding County, Sichuan Province, China and identified as B. pinchonii based on its morphometric features and DNA barcoding technology. The specimen used in this study had Animal Ethics approval for experimentation granted by Chengdu University of Traditional Chinese Medicine. The complete mitochondrial genome of B. pinchonii was 16,381 bp in size (GenBank accession number KX757918), which consisted of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a non-coding control region (D-loop). The overall base composition of the entire genome was 33.9% for A, 19.7% for C, 13.6% for G, and 32.8% for T and had low GC content of 33.3%. Except for COI, all the 13 protein-coding genes initiated with ATG as start codon. Six protein-coding genes (COI, ATP8, ATP6, ND3, ND4L, and ND5) ended with typical complete stop codon TAA, while ND1 was terminated with TAG. Other six genes (ND2, COII, COIII, ND4, ND6, and Cytb) terminated with T as the incomplete stop codons, which were presumably completed as TAA by post-transcriptional polyadenylation (Anderson et al. Citation1981). All protein-coding genes, except for ND6, were encoded on H-strand. The longest one was ND5 gene (1821 bp) in all protein-coding genes, however, the shortest was ATP8 gene (168 bp). The length of 12S rRNA and 16S rRNA genes were 934 bp and 1602 bp, respectively. The D-loop region was 793 bp in size and located between tRNA-Pro and tRNA -Phe.
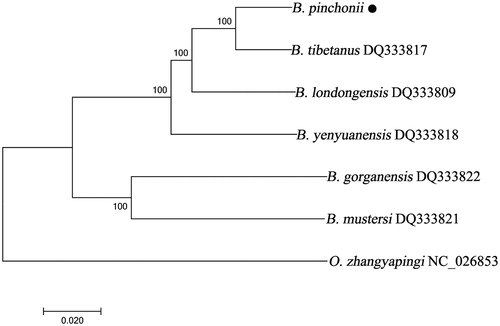
To validate the phylogenetic position of B. pinchonii, a neighbour-joining tree (with 1000 bootstrap replicates) was constructed using MEGA7 including complete mitogenomes of seven species derived from Hynobiidae. Onychodactylus zhangyapingi was used as an outgroup for tree rooting. Result showed that B. pinchonii was placed mostly close to Batrachuperus tibetanus ().
Disclosure statement
The authors declare no any conflict of interest in the preparation and execution of this manuscript.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Che J, Chen HM, Yang J, Jin JX, Jiang K, Yuan ZY, Murphy BW, Zhang YP. 2012. Universal COI primers for DNA barcoding amphibians. Mol Ecol Res. 12:247–258.
- Chen MY, Mao RL, Liang D, Kuro-o M, Zeng XM, Zhang P. 2015. A reinvestigation of phylogeny and divergence times of Hynobiidae (Amphibia, Caudata) based on 29 nuclear genes. Mol Phylogenet Evol. 83:1–6.
- Du WJ, Liu HQ, Xu J, Luo GF, Mei ZN. 2015. Molecular identification of Tibetan medicine Qianghuoyu by COI. China J Chinese Materia Medica. 40:395–398.
- Qi CH, Gong DJ, Wu HC, Hou F. 2010. Further research on Chinese Herbal Medicine Batrachuperus tibetanus. J Anhui Agri Sci. 38:9039–9040.