Abstract
We report for the first time, the complete mitochondrial genome sequence of the porbeagle shark, Lamna nasus, from a specimen collected from offshore waters of New England, USA in the western North Atlantic Ocean. The genome structure of this species is similar to the other reported shark mitogenomes. The genome sequence has a total length of 16,697 bases; similar in size to the mtDNA genomes reported for other lamnid species. A Bayesian phylogenetic tree was reconstructed for the Lamnidae family using mitogenome sequences available in the Genbank.
The porbeagle shark, Lamna nasus, is a coastal and oceanic species occurring globally within temperate and cold-temperate waters. This species is currently classified as Vulnerable by the International Union for Conservation of Nature (IUCN) Red List due to low-reproductive capacity, high commercial value and subsequent over-exploitation of discrete populations throughout its range (Stevens et al. Citation2006). Populations within both the eastern and western North Atlantic Ocean have both been significantly overfished, with a collapse of the Northwest Atlantic population in approximately 6 years (Campana et al. Citation2002). Recent stock assessments suggest that some populations have declined by up to 90% via overfishing and populations globally are considered overfished (Curtis et al. Citation2016). However, recent management improvements in some areas appear to show that populations have stabilized or are starting to rebuild (Curtis et al. Citation2016).
In this study, we report the complete mitochondrial genome of the porbeagle shark (GenBank accession number KX610464) of a specimen collected by Cape Canaveral Scientific, Inc. from New England waters off the Atlantic coast, USA. A dorsal white muscle sample was removed, frozen and the DNA processed at the University of Georgia (UGA), and subsequently stored at the Instituto de Ciencias del Mar y Limnología, UNAM. The genomic DNA was isolated using a phenol/chloroform protocol and the genomic DNA was hydrated in buffer TE. For the library preparation, the DNA was sheared by sonication with Bioruptor® (Denville, NJ) using 5 cycles of 30 seconds sonicating and 30 seconds without sonication, to obtain optimal fragment size (≤500 bp). The KAPA BIOSYSTEMS® (Wilmington, MA) Hyper Prep Kit, Illumina (San Diego, CA) was used for library preparation protocol. In brief, fragmented DNA was ligated to Illumina universal TruSeq adapters containing 8 custom nucleotide indexes (Glenn et al. Citation2016), amplification by 15 PCR cycles and library cleaning with magnetic beads. Library sequencing was performed on an Illumina®TM NextSeq using the 500/550 kit to produce paired-end 150 nucleotide reads in the Georgia Genomics Facility from UGA.
A total of 1,495,488X2 reads were obtained, quality filtered and assembled in Geneious® 8.1.7 (Auckland, New Zealand) using as reference the mtDNA genome of L. ditropis (Accession number NC_024269). The consensus sequence obtained was annotated using the software MitoAnnotator (Iwasaki et al. Citation2013).
The assembled mitogenome has a total length of 16,697 bases, containing 13 protein-coding genes, 2 rRNA genes, 22 tRNAs, and the control region. The overall base composition of the genome was: A: 29.7%, T: 28.3%, C: 27.2% and G: 14.7%.
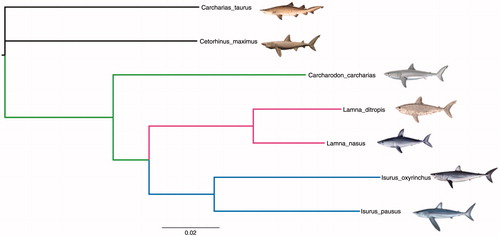
The genome sequence of the porbeagle shark was aligned against the complete mitochondrion genomes reported for all the other four living species of the Lamnidae family: the white shark, Carcharodon carcharias, longfin mako shark, Isurus paucus, shortfin mako shark, I. oxyrinchus, and salmon shark, Lamna ditropis (). A phylogenetic tree was constructed including sequences of basking shark, Cetorhinus maximus, and sand tiger shark, Carcharias taurus, as out-groups, using MrBayes (Ronquist & Huelsenbeck Citation2003). About 10,000,000 generations were used for length chain, and 100,000 for burn-in length. The Lamnidae relationships agree in grouping Carcharodon (Isurus + Lamna) (), which also coincides with the parsimony phylogeny obtained for the mitochondrial gene ND2 using transitions and transversions in the analyses by Naylor et al (Citation1997), and the phylogeny obtained for Lamnidae with whole mtDNA sequences in Díaz-Jaimes et al. (Citation2016). Nevertheless, it differs from the phylogeny Lamna (Carcharodon + Isurus) obtained with morphological and other molecular data to date (Compagno Citation1990; Vélez-Zuazo & Agnarsson Citation2011). This result should be considered with caution as it represents the phylogeny of the whole mitogenome molecule and not necessarily the group phylogeny.
Figure 1. Bayesian phylogenetic tree from mitogenome sequences for the five species of the family Lamnidae: Carcharodon carcharias (NC_022415), Isurus paucus (NC_024101), Isurus oxyrinchus (NC_022691), Lamna ditropis (NC_024269), Lamna nasus (KX610464), and Cetorhinus maximus (NC_023266) and Carcharias taurus (NC_023520) as outgroup. All posterior probabilities were equal to 1.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Campana SE, Joyce W, Marks L, Natanson LJ, Kohler NE, Jensen CF, Mello JJ, Pratt HL, Jr., Myklevoll S. 2002. Population dynamics of the porbeagle in the Northwest Atlantic Ocean. North Am J Fish Manage. 22:106–121.
- Compagno LJV. 1990. Relationships of the megamouth shark, Megachasma pelagios (Lamniformes: Megachasmidae), with comments on its feeding habits. NOAA Technical Report NMFS, vol. 90, p. 357–379
- Curtis TH, Laporte S, Cortes E, DuBeck G, McCandless C. 2016. Status review report: Porbeagle Shark (Lamna nasus). Final Report to National Marine Fisheries Service, Office of Protected Resources. February 2016. 56 pp.
- Díaz-Jaimes P, Bayona-Vásquez NJ, Adams DH, Uribe-Alcocer M. 2016. Complete mitochondrial DNA genome of bonnethead shark, Sphyrna tiburo, and phylogenetic relationships among main superorders of modern elasmobranchs. Meta Gene. 7:48–55.
- Glenn TC, Nilsen R, Kieran TJ, Finger JW, Pierson TW, Bentley KE, Hoffberg S, Louha S, Garcia-De-Leon FJ, Angel del Rio Portilla M, et al. 2016. Adapterama I: universal stubs and primers for thousands of dual-indexed Illumina libraries (iTru & iNext). BioRxiv. http://dx.doi.org/10.1101/049114.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh KP, Tetsuya S, Mabuchi K, Hirohiko T, Miya M, Nishida M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Naylor GJP, Martin AP, Mattison EG, Brown WM. 1997. Interrelationships of lamniform sharks: testing phylogenetic hypotheses with sequence data. In: Kocher TD, Stepien CA. editors. Molecular systematics of fishes San Diego: Academic Press; p. 199–218.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 20:407–415.
- Stevens J, Fowler SL, Soldo A, McCord M, Baum J, Acuña E, Domingo A, Francis M [Internet]. 2006. Lamna nasus. The IUCN Red List of Threatened Species 2006: e.T11200A3261697. Available from: http://dx.doi.org/10.2305/IUCN.UK.2006.RLTS.T11200A3261697.en.
- Vélez-Zuazo X, Agnarsson I. 2011. Shark tales: a molecular phylogeny of sharks (Selachimorpha, Chondrichtyes). Mol Phylogenet Evol. 58:207–217.
