Abstract
In this study, we determined the complete plastome sequence of Carissa macrocarpa (Eckl.) A. DC. (Apocynaceae) (NCBI acc. no. KX364402). The gene order and structure of the C. macrocarpa plastome are similar to those of a typical angiosperm. The complete plastome is 155,297 bp in length, and consists of a large single-copy region of 85,586 bp and a small single-copy region of 18,131 bp, which are separated by two inverted repeats of 25,792 bp. The plastome contains 113 genes, of which 79 are protein-coding genes, 30 are tRNA genes and 4 are rRNA genes. Sixteen genes contained one intron and two genes have two introns. The average A–T content of the plastome is 62.0%. A total of 31 simple sequence repeat loci were identified within the genome. Phylogenetic analysis revealed that C. macrocarpa is a member of the paraphyletic subfamily Rauvolfioideae of Apocynaceae. The sister group relationship of C. macrocarpa to the Apocynoideae–Asclepiadoideae clade is supported by 100% bootstrap values.
Carissa macrocarpa (Eckl.) A. DC., one of the tropical fruits of the family Apocynaceae, is native to South Africa (Kim Citation2011). This species is cultivated as an ornamental and edible plant. C. macrocarpa belongs to the subfamily Rauvolfioideae of the family Apocynaceae, which is one of the five families in the order Gentianales (APG IV Citation2016). In terms of species number, Apocynaceae is the 10th largest family of flowering plants. The family consists of 5 subfamilies, 366 genera and 5100 species (Christenhusz & Byng Citation2016), with most species being distributed in the subtropical and tropical regions. The family includes many economic plants, such as species of Catharanthus, Carissa, and Nerium, which are important sources of medicine and fruits. To date plastome sequences have been determined for 10 species within the Apocynaceae (Ku et al. Citation2013; Straub et al. Citation2013, Citation2014; Park et al. Citation2014; Jang et al. Citation2015; Park et al. Citation2016). Our data for C. macrocarpa will be the first complete plastome sequence for a species in the genus Carrisa and tribe Carisseae.
The leaves of C. macrocarpa used in this study were collected from the Korea University greenhouse, where we grew the plants from seeds originally collected from South Africa. The plants flowered and fruited in the greenhouse. A voucher specimen was deposited in the Korea University Herbarium (KUS acc. no. 2014-0239). Fresh leaves were ground into powder in liquid nitrogen and total DNAs were extracted using the CTAB method (Doyle & Doyle Citation1987). The DNAs were further purified by the ultracentrifugation and dialysis (Palmer Citation1986). The genomic DNAs are deposited in the Plant DNA Bank in Korea (PDBK acc. no. 2014-0239). The complete plastome sequence was generated using an Illumina HiSeq 2000 system (Illumina Inc., San Diego, CA). Annotations were performed using the National Center for Biotechnology Information (NCBI) BLAST, DOGMA (Wyman et al. Citation2004), and tRNAscan-SE programs (Lowe & Eddy Citation1997). For the phylogenetic analysis, we selected and downloaded 38 complete plastome sequences based on the APG IV system (APG IV Citation2016) from the NCBI database.
The gene order and structure of the C. macrocarpa plastome are similar to those of a typical angiosperm (Shinozaki et al. Citation1986; Kim & Lee Citation2004; Yi & Kim Citation2012). The complete plastome is 155,297 bp in length and consists of a large single-copy (LSC) region of 85,582 bp and a small single-copy (SSC) region of 18,129 bp, which are separated by two inverted repeats (IR) of 25,793 bp. The plastome comprises 113 unique genes (79 protein-coding genes, 30 tRNA genes and 4 rRNA genes). The average A–T content of the plastome is 62.0%. The A–T contents in the LSC, SSC and IR regions are 63.9%, 67.9%, and 56.7%, respectively. The average coverage of the sequence is 1,057×. Sixteen genes contain one intron and two genes, ycf3 and clpP, have two introns. A total of 31 simple sequence repeat (SSR) loci are distributed throughout the plastome. Among these, 23, 3 and 5 are mono-SSR, di-SSRand tri-SSR loci, respectively. Some of these loci will be useful in identifying cultivars of C. macrocarpa.
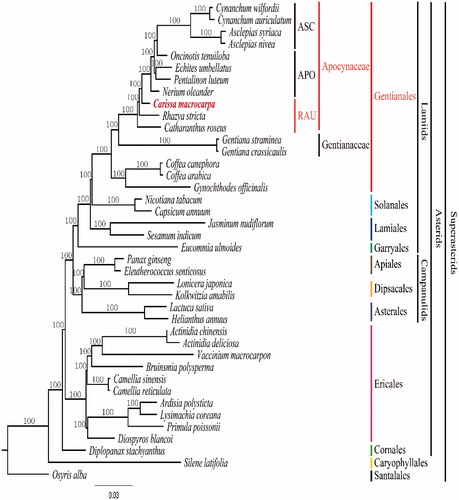
To validate the phylogenetic relationships of C. macrocarpa among Apocynaceae, we constructed a maximum likelihood tree. Phylogenetic analysis was performed on a data set that included 79 protein-coding genes and 4 rRNA genes from 40 taxa using RAxML v. 7.7.1 (Stamatakis et al. Citation2008). The 83 gene sequences (86,114 bp) were aligned with MUSCLE in Geneious v. 6.1.8 (Biomatters Ltd.; Kearse et al. Citation2012). The sister group relationship of C. macrocarpa to the Apocynoideae-Asclepiadoideae clade is supported by 100% bootstrap values (). The monophyly of subfamily Apocynoideae is supported in the plastome tree, whereas the paraphylies of Asclepiadoideae and Rauvolfioideae are suggested. Our tree based on complete plastome data, has a similar topology to those generated in previous phylogenetic studies based on more limited gene sequence data (Potgieter & Albert Citation2001; Sennblad & Bremer Citation2002; Simões et al. Citation2007). In order to clarify the phylogenetic relationships more precisely, we need additional complete plastome data from two other poorly studied subfamilies, Periplocoideae and Secamonoideae.
Figure 1. Maximum Likelihood (ML) tree based on 79 protein-coding and 4 rRNA genes from 40 plstomes as determined by RAxML(−ln L = −527039.471210). The numbers at each node indicate the ML bootstrap values. Three subfamilies within Apocynaceae are abbreviated as follows: APO: Apocynoideae; ASC: Asclepiadoideae; RAU: Rauvolfioideae. Genbank accession numbers of taxa are shown below, Actinidia chinensis (NC_026690), Actinidia deliciosa (NC_026691), Ardisia polysticta (NC_021121), Asclepias nivea (NC_022431), Asclepias syriaca (NC_022432), Bruinsmia polysperma (NC_030180), Camellia reticulata (NC_024663), Camellia sinensis (NC_020019), Capsicum annuum (NC_018552), Carissa macrocarpa (KX364402), Catharanthus roseus (NC_021423), Coffea arabica (NC_008535), Coffea canephora (NC_030053), Cynanchum auriculatum (NC_029460), Cynanchum wilfordii (NC_029459), Diospyros blancoi (KX426216), Diplopanax stachyanthus (NC_029750), Echites umbellatus (NC_025655), Eleutherococcus senticosus (NC_016430), Eucomia ulmoides (KU204775), Gentiana crassicaulis (NC_027442), Gentiana straminea (NC_027441), Gynochthodes offcinalis (NC_028009), Helianthus annuus (NC_007977), Jasminum nudiflorum (NC_008407), Kolkwitzia amabilis (NC_029874), Lactuca sativa (NC_007578), Lonicera japonica (NC_026839), Lysimachia coreana (NC_026197), Nerium oleander (NC_025656), Nicotiana tabacum (NC_001879), Oncinotis tenuiloba (NC_025657), Osyris alba (NC_027960), Panax ginseng (NC_006290), Pentalinon luteum (NC_025658), Primula poissonii (NC_024543), Rhazya stricta (NC_024292), Sesamum indicum (NC_016433), Silene latifolia (NC_016730), and Vaccinium macrocarpon (NC_019616).
Disclosure statement
The authors report no conflicts of interest, and are independently responsible for the content and writing of the paper.
Additional information
Funding
References
- APG IV. 2016. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Christenhusz MJ, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa. 261:201–217.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jang W, Kim K-Y, Kim K, Lee S-C, Park H-S, Lee J, Seong RS, Shim YH, Sung SH, Yang T-J. 2015. The complete chloroplast genome sequence of Cynanchum auriculatum Royle ex Wight (Apocynaceae). Mitochondrial DNA. 10.3109/19401736.2015.1101557
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim K-J, Lee HL. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11:247–261.
- Kim K-J. 2011. Tropical fruit resources. Seoul: Geobook.
- Ku C, Chung W-C, Chen L-L, Kuo C-H. 2013. The complete plastid genome sequence of Madagascar periwinkle Catharanthus roseus (L.) G. Don: plastid genome evolution, molecular marker identification, and phylogenetic implications in asterids. PLoS One. 8:e68518.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Palmer JD. 1986. Isolation and structural analysis of chloroplast DNA. Method Enzymol. 118:167–186.
- Park H-S, Kim K-Y, Kim K, Lee S-C, Lee J, Seong RS, Shim YH, Sung SH, Yang T-J. 2016. The complete chloroplast genome sequence of an important medicinal plant Cynanchum wilfordii (Maxim.) Hemsl. (Apocynaceae). Mitochondrial DNA Pt A. 27:3747–3748.
- Park S, Ruhlman TA, Sabir JS, Mutwakil MH, Baeshen MN, Sabir MJ, Baeshen NA, Jansen RK. 2014. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics. 15:1–18.
- Potgieter K, Albert VA. 2001. Phylogenetic relationships within Apocynaceae sl based on trnL intron and trnL-F spacer sequences and propagule characters. Ann Missouri Bot Gard. 88:523–549.
- Sennblad B, Bremer B. 2002. Classification of Apocynaceae s.l. according to a new approach combining Linnaean and phylogenetic taxonomy. Syst Biol. 51:389–409.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. Embo J. 5:2043–2049.
- Simões AO, Livshultz T, Conti E, Endress ME. 2007. Phylogeny and systematics of the Rauvolfioideae (Apocynaceae) based on molecular and morphological evidence 1. Ann Missouri Bot Gard. 94:268–297.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Straub SC, Cronn RC, Edwards C, Fishbein M, Liston A. 2013. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae). Genome Biol Evol. 5:1872–1885.
- Straub SC, Moore MJ, Soltis PS, Soltis DE, Liston A, Livshultz T. 2014. Phylogenetic signal detection from an ancient rapid radiation: effects of noise reduction, long-branch attraction, and model selection in crown clade Apocynaceae. Mol Phylogenet Evol. 80:169–185.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yi D-K, Kim K-J. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One. 7:e35872.
