Abstract
The complete mitochondrial genome of Typhlatya miravetensis from one of its only three known localities (Ullal de la Rabla de Miravet, Castellón, Spain) is presented here. The mitogenome is 15,865 bp in length and includes the standard set of two rRNAs, two non-coding regions plus 13 protein-coding genes. The later have been used to perform a phylogenetic analysis together with other Caridea representatives with mitogenome data in GenBank, inferring a close relationship with the Hawaiian volcano shirmp (Halocaridina rubra) within the family Atyidae.
Typhlatya Creaser, 1936 is an obligate subterranean aquatic genus of atyid shrimps that comprises 17 species, showing a punctuated distribution throughout coastal continental and insular ground-waters of the Mediterranean, north-central Atlantic and east Pacific (Botello et al. Citation2013). Such peculiar biology and distribution have classically drawn the attention of evolutionary biologists (e.g. Croizat et al. Citation1974; Banarescu Citation1990; Stock Citation1993; Hunter et al. Citation2008; Botello et al. Citation2013). Although a few of the studies are based on the analysis of DNA sequences, our knowledge about the genetics of this group of organisms is still scarce. Here we present the first mitogenome sequence for the genus Typhlatya. The T. miravetensis Sanz & Platvoet Citation1995 sample was collected in 2009 in Ullal de la Rambla de Miravet (Castellón, Spain; 40° 06’ 73’’N, 00° 03’ 60’’W). Voucher specimen has been deposited at the DNA and tissue collection of the Biodiversity, Systematic and Evolution group (Bio6Evo) of the University of the Balearic Islands with accession number 3151.
Total DNA was purified and the mitogenome was amplified in two large amplicons through long-range PCR as described in Pons et al. (Citation2014). Amplicons were sequenced using a Junior NGS 454 platform. Genes were annotated with DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013) webservers. A complete mitogenome of 15,865 bp was obtained (ENA accession number LT608343).
The mitochondrial genome shows an A + T bias (A + T = 66.28%) and the gene content including 13 protein-coding genes (PCGs), 22 tRNAs and two ribosomal genes. An AT-rich (79.94%) non-coding region of 1042 bp was found between rrnS and trnI genes and might correspond to the origin of replication. Gene arrangement matches the ancestral putative pancrustacean gene order. Most genes are encoded in the positive strand excepting nad1, nad4, nad4L, nad5, rrnL, rrnS and the tRNAs L1, C, F, H, P, Q, V and Y. Start codons match the canonical ATA or ATG except for cox1 (CCG), nad5 (GTG), nad6 (ATC), atp8, nad1 and nad2 (ATT). Similarly, stop codons are the canonical TAA or TAG except for cox2 that showed a truncated codon stop (T).
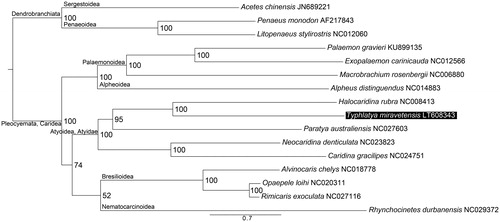
Representative species from all Caridea genera with mitogenome sequences available in GenBank plus three Dendrobranchiata taxa as outgroup were used to assess the phylogenetic placement of T. miravetensis, which showed a close relationship with Halocaridina rubra Holthuis, 1963 within the family Atyidae ().
Figure 1. Maximum likelihood tree obtained using IQTREE v 1.3.12 (Nguyen et al. Citation2015) based on the PCGs of 12 Caridea species and three Dendrobranchiata outgroups highlighting the phylogenetic placement of T. miravetensis. DNA sequences were aligned in TranslatorX (Abascal et al. Citation2010). PartitionFinder v 1.1.1 (Lanfear et al. Citation2012) was used to select the best partitioning scheme and evolutionary models. Node numbers show bootstrap support values. GenBank accession numbers are given with species names.
Acknowledgements
We thank Alberto Sendra (Valencia) who collected the specimen used in this study.
Disclosure statement
The authors report no conflicts of interest and are responsible for the content and writing of the article.
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–13.
- Banarescu P. 1990. Zoogeography of fresh waters. In General Distribution and Dispersal of Freshwater Animals Vol. 1. Wiesbaden, Germany: Aula-Verlag.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Botello A, Iliffe TM, Alvarez F, Juan C, Pons J, Jaume D. 2013. Historical biogeography and phylogeny of Typhlatya cave shrimps (Decapoda: Atyidae) based on mitochondrial and nuclear data. J Biogeogr. 40:594–607.
- Croizat L, Nelson G, Rosen DE. 1974. Centers of origin and related concepts. Syst Zool. 23:265.
- Hunter RL, Webb MS, Iliffe TM, Bremer JRA. 2008. Phylogeny and historical biogeography of the cave-adapted shrimp genus Typhlatya (Atyidae) in the Caribbean sea and western Atlantic. J Biogeogr. 35:65–75.
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Pons J, Bauzà-Ribot MM, Jaume D, Juan C, Waeschenbach A, Telford M, Porter J, Littlewood D, Fernández-Silva P, Enriquez J, et al. 2014. Next-generation sequencing, phylogenetic signal and comparative mitogenomic analyses in Metacrangonyctidae (Amphipoda: Crustacea). BMC Genom. 15:566.
- Sanz S, Platvoet D. 1995. New perspectives on the evolution of the genus Typhlatya (Crustacea, Decapoda): first record of a cavernicolous atyid in the Iberian Peninsula, Typhlatya miravetensis n. sp.. Contr. Zool. 65:79–99.
- Stock JH. 1993. Some remarkable distribution patterns in stygobiont Amphipoda. J Nat Hist. 27:807–819.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
