Abstract
Hydrocotyle sibthorpioides Lam. is a widespread and important Chinese medicinal plant. In this study, we have sequenced the complete chloroplast genome of H. sibthorpioides, which is 152,880 bp in length with large (LSC 84,064 bp) and small (SSC 18,690 bp) single-copy regions separated by a pair of inverted repeats (IRs 25,063 bp) and contains 113 unique genes with 17 genes duplicated in the IR making a total of 130 genes. The phylogenetic analysis strongly supports the position of H. sibthorpioides and indicates it belongs to the Araliaceae family, potential to facilitate a better understanding of population and phylogenetic studies in Apiales.
Hydrocotyle sibthorpioides is a widespread and perennial herb. As an important Chinese medicinal plant, it is often used for treating immune disorders and liver diseases. It is reported that it is effective in the treatment of rheumatalgia, dysentery, jaundice, and exerted a potent inhibitory effect on the growth of tumours (Huang et al. Citation2013).
Hydrocotyle sibthorpioides used to belong to Apiaceae, but in mostly molecular-based APG III system (APG, Citation2009), its genus, Hydrocotyle, is placed in Araliaceae. Considering that chloroplast DNA (cpDNA) conservativity and its slow rate of nucleotide substitution make cpDNA widely used in plant phylogeny (Olmstead & Palmer Citation1994); thus, we report complete chloroplast genome of H. sibthorpioides and conduct phylogenetic trees for intention of confirming its position in Apiales.
Fresh leaves of H. sibthorpioides were collected from Zhejiang University. Voucher specimen (accession number: CJ201505140311a) were deposited into Institute of Biomedical Engineering of Zhejiang University, Hangzhou, China. Pure cpDNA was sequenced using Illumina sequencing platform (Illumina Inc., San Diego, CA). Illumina sequencing data were assembled using SOAPdenovo (Luo et al. Citation2012). The chloroplast genome was annotated using DOGMA (Wyman et al. Citation2004) with manual correction. After using Gblocks Version 0.91b (Talavera & Castresana Citation2007) for the selection of conserved regions and Modeltest version 3.7 (Posada & Crandall Citation1998) for appropriate model selection, maximum likelihood (ML) analysis were performed using RAxML 8.0.19 (Stamatakis Citation2014).
The complete chloroplast genome of H. sibthorpioides (GenBank assession number KT589392) is 152,880 bp in length with 62.49% AT content, and shows a typical quadripartite structure with a pair of inverted repeats (IRs, 25,063 bp), separated by a small single-copy region (SSC, 18,690 bp) and a large single copy region (LSC, 84,064 bp), respectively. This chloroplast genome contains 113 unique genes consisting of 79 protein-coding genes, four ribosomal RNA genes and 30 transfer RNA genes, with 17 genes duplicated in the IR making a total of 130 genes, and ycf1, ycf15, rps19 in the IR regions are pseudogene. The gene organization and content of H. sibthorpioides are similar to those previously reported species belonging to the Araliaceae family, especially Hydrocotyle verticillata.
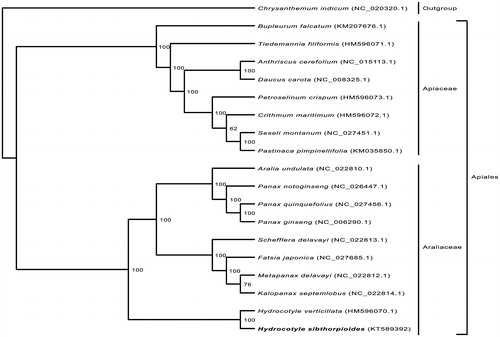
The phylogenetic relationship of H. sibthorpioides was deduced by its comparison with other 17 Apiales chloroplast genomes and Chrysanthemum indicum added as the outgroup based on 80 common protein-coding genes. ML analyses yielded a topology structure with high bootstrap support values (). In ML tree, there are two families, Araliaceae and Apiaceae, in Apiales. The chosen core Araliaceae strongly formed two major groups consisting with previous studies (Li et al. Citation2013). As placed in Araliaceae family, the Hydrocotyle genus is the earliest diverging lineage, which was identified as sister to the rest with strong support. Despite the fact that H. sibthorpioides used to belong to Apiaceae in traditional plant classification such as Cronquist system, this result is consistent with previous phylogenetic studies (Lowry et al. Citation2001; Plunkett et al. Citation2004; Nicolas & Plunkett Citation2009; Chen et al. Citation2016) and APG III system.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the Jiangsu Technology Support Program under Grant No. BE2014654, Jiangsu Provincial Natural Science Foundation of China under Grant No. BK20161269, and Changshu Technology Support Program under Grant No. CS201405.
References
- APG. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linnean Soc. 161:105–121.
- Chen Q, Feng X, Li M, Yang B, Gao C, Zhang L, Tian J. 2016. The complete chloroplast genome sequence of Fatsia japonica (Apiales: Araliaceae) and the phylogenetic analysis. Mitochondrial DNA. 27:3050–3051.
- Huang Q, Huang R, Zhang S, Lin J, Wei L, He M, Zhuo L, Lin X. 2013. Protective effect of genistein isolated from Hydrocotyle sibthorpioides on hepatic injury and fibrosis induced by chronic alcohol in rats. Toxicol Lett. 217:102–110.
- Li R, Ma P-F, Wen J, Yi T-S. 2013. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLoS One. 8:e78568.
- Lowry P II, Plunkett G, Oskolski A. 2001. Early lineages in Apiales: insights from morphology, wood anatomy and molecular data. Edinburgh J Bot. 58:207–220.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Nicolas AN, Plunkett GM. 2009. The demise of subfamily Hydrocotyloideae (Apiaceae) and the re-alignment of its genera across the entire order Apiales. Mol Phylogenet Evol. 53:134–151.
- Olmstead RG, Palmer JD. 1994. Chloroplast DNA systematics: a review of methods and data analysis. Am J Bot. 81:1205–1224.
- Plunkett GM, Chandler GT, Lowry PP, Pinney SM, Sprenkle TS, van Wyk B-E, Tilney P. 2004. Recent advances in understanding Apiales and a revised classification. South Afr J Bot. 70:371–381.
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.