Abstract
The mitogenome sequence of a polychaete Hediste diadroma (Phyllodocida, Nereididae) was determined first in the genus Hediste. The circular genome is 15,765bp in size including 13 protein-coding genes (PCGs), 2 rRNA and 22 tRNA of which the order and structure is same as those of other Nereididae species. All PCGs use ATG as the start codon while for the stop codon, COI, ND5 and ND1 use an incomplete codon of T. The genome consists of 32.4% A, 18.2% C, 14.2% G, 35.2% T, showing a high content of A + T similar to the other Phyllodocid polychaetes.
The genus Hediste (Malmgren Citation1867) contains only five species which occur mostly in macro-benthic fauna of shallow brackish water, being distributed along the East Asia(Sato & Nakashima Citation2003), the Atlantic coasts of Europe and the North America (Smith Citation1977), and the North American Pacific coast (Smith Citation1958). Hediste diadroma is often a dominant mudflat species in the Korean coast of the Yellow sea and in the Japanese estuaries (Sato & Nakashima Citation2003). In this study, we determined the complete mitochondrial genome of H. diadroma, first in the genus Hediste.
Specimens of H. diadroma collected from the subtidal zone of the Masan harbour in the southern coast of Korea. The voucher specimens are deposited in National Marine Biodiversity Institute of Korea (MABIK NA00143000-00143036). The genomic DNA was extracted from the muscle tissue and the mitogenome sequences were analyzed in two ways: First, dozens of mitochondrial DNA fragments were amplified through PCR with primer sets designed from the partial sequences of COI, COIII, CytB, ND4 and 16S genes of H. diadroma, and sequenced by the Sanger method, which resulted in a sequence of 12,578bp; Then, the gap sequence of 3178bp was filled up by application of Illumina Hiseq2000 sequencing platform. The sequences were assembled and annotated in comparison with the previously reported mitogenome sequences of Nereididae species (Boore Citation2001; Kim et al. Citation2015) usingGeneious v9.1.2 (Biomatters Ltd. Auckland, New Zealand; Kearse et al. Citation2012).
The circular mitogenome of H. diadroma (GeneBank accession number KX499500) is 15,765bp in length, which includes 2 rRNA, 22 tRNA, and 13 protein-coding genes (PCGs) as well as the control region. All genes are encoded on the H-strand and their order and structure in the genome is identical to those of other Nereididae species. The overall nucleotide composition is 32.4% A, 18.2% C, 14.2% G, 35.2% T, showing a high A + T bias (67.6%). All PCGs get off by the typical ATG start codon. Nine of 13 PCGs use TAA for the stop codon, and the ND6 gene ends with TAG while COI, ND5 and ND1 genes have an incomplete stop codon, T. The lengths of tRNA genes range from 54 to 67bp and all tRNAs except tRANArg, tRNASerUCN, and tRNASerAGN have the typical clover leaf structure. The three tRNAs has a reduced DHU arm. The control region is 1093bp long, being located between tRNAGly and tRNATyr. The sizes of 12S and 16S rRNAs are 803bp and 1196bp, respectively.
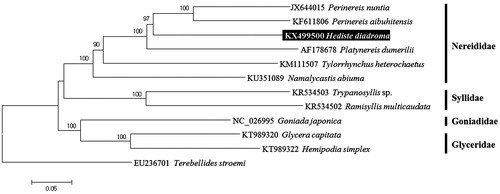
The taxonomic position of H. diadroma analyzed by molecular phylogeny of the mitogenome using the neighbour-joining (NJ) method with the K2P model in MEGA 6 (Tamura et al. Citation2013) reveals that H. diadroma is closely related with Perinereis spp. Their cluster is strongly supported by bootstrap values of more than 97% (). The mitogenome of H. diadroma, the first reported of its genus will provide useful information for understanding of evolutionary history and phylogeny of the genus Hediste in relation to the other genera within the family Nereididae.
Figure 1. Neighbour-joining tree for six species of the family Nereididae including Hediste diadroma and five other related species in Phyllodocida inferred from mitochondrial genomes. Terebellides stroemi (EU236701) derived from Terebellida was used as outgroup for tree rooting. Data set for Phylogenetic trees were used nucleotide sequences of 13 protein-coding genes. Numbers above the branches indicate NJ bootstrap values from 1000 replication.
Disclosure statement
The authors declare that they do not have any conflict of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the grants from National Marine Biodiversity Institute of Korea (2016M00300) and Korea Institute of Ocean Science and Technology.
References
- Boore JL. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Mol Biol Evol. 18:1413–1416.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Thierer T. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim H, Jung G, Lee Y-C, Pae SJ, Kim CG, Lee Y-H. 2015. The complete mitochondrial genome of the marine polychaete: Perinereis aibuhitensis (Phyllodocida, Nereididae). Mitochondrial DNA. 26:869–870.
- Malmgren AJ. 1867. Annulata Polychaeta: Spetsbergiae, Groenlandiae, Islandiae et Scandinaviae. Hactenus Cognita. Ex Officina Frenckelliana; p. 48.
- Sato M, Nakashima A. 2003. A review of Asian Hediste species complex (Nereididae, Polychaeta) with descriptions of two new species and a redescription of Hediste japonica (Izuka, 1908). Zool J Linnean Soc. 137:403–445.
- Smith RI. 1958. On reproductive pattern as a specific characteristic among nereid polychaetes. Syst Biol. 7:60–73.
- Smith RI. 1977. Physiological and reproductive adaptations of Nereis diversicolor to life in the Baltic Sea and adjacent waters. Essays on Polychaetous Annelids. In Memory of Dr. Olga Hartman. Los Angeles (CA): University of California; p. 373–390.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2725–2729.
