Abstract
The Korean oily shinner (Sarcocheilichthys nigripinnis morii) is a subspecies belonging to the subfamily Gobioninae in the family Cyprinidae and is endemic to Korea. Herein, we report the first sequencing and assembly of the complete mitochondrial genome of S. nigripinnis morii. The complete mitochondrial genome is 16,679 bp long, consisting of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. It has the typical vertebrate mitochondrial gene arrangement. Phylogenetic analysis using mitogenomes of 16 species showed that S. nigripinnis morii was clustered with S. nigripinnis and S. variegatus microoculus and grouped with the other Cyprinidae species.
The Korean oily shinner, Sarcocheilichthys nigripinnis morii (Jordan & Hubbs Citation1925), is a subspecies belonging to the subfamily Gobioninae in the family Cyprinidae and is endemic to Korea. This subspecies is allopatrically distributed in upstream and midstream regions in southwestern Korea (Kim & Park Citation2007). To date, there has been very little molecular and genetic research on this species. To the best of our knowledge, this is the first study to determine the complete mitochondrial genome of S. nigripinnis morii and to analyze the phylogenetic relationship of this subspecies among Cypriniformes fishes.
The S. nigripinnis morii specimen (standard length, 93.6 mm) was collected from the Mangyeong River, South Korea (35.47°N, 127.11°E). The specimen was deposited in the National Institute of Biological Resources (NIBR, Voucher No. NIBRGR0000098804). Genomic DNA from muscle tissue was sequenced and assembled using the Illumina Hiseq 4000 sequencing platform (Illumina, San Diego, CA) and SOAPdenovo assembler at Macrogen Inc. (Korea), respectively. The complete mitochondrial genome was annotated using MacClade ver. 4.08 (Maddison & Maddison Citation2005) and DNASIS ver. 3.2 (Hitachi Software Engineering). Experiments were conducted in accordance with the Guidelines of Animal Ethics published by the NIBR.
The complete mitochondrial genome of S. nigripinnis morii (GenBank accession no. AP017653) is 16,679 bp long and includes 13 protein-coding, 22 tRNA, and 2 rRNA genes. The ND6 gene and eight tRNA genes are encoded on the light strand. The overall base composition of the heavy strand is 29.83% A, 26.70% C, 17.21% G, and 26.24% T. As in mitogenomes of other vertebrates (Saccone et al. Citation1999), the AT content is higher than the GC content. All tRNA genes can fold into a typical cloverleaf structure and are 68–76 bp long. The 12S rRNA (958 bp) and 16S rRNA genes (1690 bp) are located between tRNAPhe and tRNAVal and between rRNAVal and tRNALeu(UUR), respectively. Except for COI, which starts with GTG, the 12 protein-coding genes start with ATG. Seven of these terminate with incomplete stop codons, T (COII, COIII, and Cytb) and TA (ND2, ATP6, ND3, and ND4), whereas the remaining six genes ended with complete stop codons (TAA or TAG). A control region (1,007 bp) is located between tRNAPro and tRNAPhe.
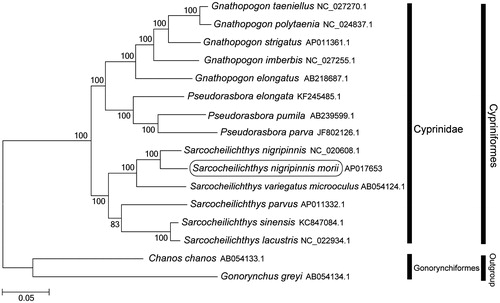
Phylogenetic trees were constructed (maximum likelihood) with 1000 replicates using MEGA 7.0 software (PA, USA) (Kumar et al. Citation2016) for the newly sequenced mitogenome and a further 15 complete mitogenome sequences downloaded from the National Center for Biotechnology Information. We confirmed that S. nigripinnis morii is clustered with S. nigripinnis (Tang et al. Citation2013) and S. variegatus microoculus (Saitoh et al. Citation2003) and grouped with the other Cyprinidae species (). Because S. nigripinnis morii exhibits a unique spawning symbiosis with mussels such as Anodonta arcaeformis (Kim & Park Citation2007; An et al. Citation2016), it is vulnerable to habitat degradation. We initiated a project to conserve them through the use of interspecies spermatogonial transplantation (Lee et al. Citation2013; Lee et al. Citation2015). This mitogenome provides a potentially important resource for developing conservation strategies and addressing taxonomic issues.
Figure 1. Phylogenetic position of Sarcocheilichthys nigripinnis morii based on a comparison with the complete mitochondrial genome sequences of 13 Cyprinidae species and 2 outgroup species. The analysis was performed using the MEGA 7.0 software. The accession numbers for each species are indicated after the scientific names.
Disclosure statement
The authors report no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Funding
This study was supported by the National Institute of Biological Resources under grant number NIBR201612107 and by the National Marine Biodiversity Institute Research Program under grant number 2016M00500.
References
- An C, Ouyang S, Zhou CH, Wu XP. 2016. The complete F-type mitochondrial genome of Chinese Anodonta arcaeformis (Bivalvia: Unionidae: Anodontinae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1552–1553.
- Jordan DS, Hubbs CL. 1925. Record of fishes obtained by David Starr Jordan in Japan. Mem Carneg Mus. 10:93–346.
- Kim IS, Park JY. 2007. Freshwater fishes of Korea. Seoul, Korea: Kyo-Hak Publishing.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee S, Iwasaki Y, Shikina S, Yoshizaki G. 2013. Generation of functional eggs and sperm from cryopreserved whole testes. Proc Natl Acad Sci USA. 110:1640–1645.
- Lee S, Seki S, Katayama N, Yoshizaki G. 2015. Production of viable trout offspring derived from frozen whole fish. Sci Rep. 5:16045.
- Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution. ver. 4.08. Sunderland (MA): Sinauer Associates.
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209.
- Saitoh K, Miya M, Inoue JG, Ishiguro NB, Nishida M. 2003. Mitochondrial genomics of ostariophysan fishes: perspectives on phylogeny and biogeography. J Mol Evol. 56:464–472.
- Tang WR, Lin HD, Su LW, Tang WQ, Wu CY, Yang JQ. 2013. The complete mitochondrial genome sequence of Sarcocheilichthys nigripinnis (Cypriniformes, Cyprinidae). Mitochondrial DNA. 24:478–480.
