Abstract
Arini with 19 genera is the most diversified tribe of the neotropical parrots from Arinae subfamily. So far, among this tribe, the genus Aratinga appeared to be the most problematic for the taxonomists. The typical representative of this genus is Aratinga solstitialis, whose complete mitochondrial genome were sequenced and compared with five other representatives of Arini mitogenomes. Despite the conservatism in their general organization, some changes in A + T% composition of individual genes, start/stop codon usage and intergenic regions accumulated during evolution.
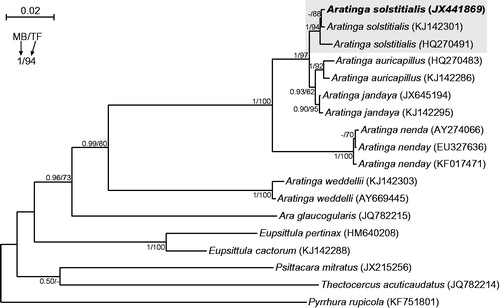
All neotropical parrots are classified into Arinae subfamily, which is divided into four tribes (Joseph et al. Citation2012; Schodde et al. Citation2013). The most taxon-rich tribe Arini includes morphologically diverse Aratinga genus, which is the most taxonomically controversial. After revision, it was split into: Eupsittula, Psittacara, Thectocercus, and ‘true’ Aratinga (Remsen et al. Citation2013; Urantowka et al. Citation2013). Here, we report the first mitogenome of Aratinga, Aratinga solstitialis (GenBank accession no. JX441869) and compared it with other Arini representatives: Ara glaucogularis (Urantowka Citation2016), Eupsittula pertinax (Pacheco et al. Citation2011), Psittacara mitratus (Urantowka et al. Citation2016a), Pyrrhura rupicola (Urantowka et al. Citation2016b), and Thectocercus acuticaudatus (Urantowka et al. Citation2013). A phylogenetic analysis confirmed undoubtedly that the obtained mtDNA belongs to Aratinga solstitialis ().
Figure 1. The phylogenetic tree obtained in MrBayes for nd2 gene indicating that the studied individual (bolded) belongs to Aratinga solstitialis. The parrot is kept in culture and its blood sample from which DNA was isolated is available in the laboratory at the Department of Genetics in Wroclaw University of Environmental and Life Sciences under the number ASS16984. Values at nodes, in the order shown, indicate posterior probabilities found in MrBayes (PP) and bootstrap percentages calculated in TreeFinder (BP). In the MrBayes (Ronquist et al. Citation2012) analysis, separate mixed substitution models were assumed for three codon positions with information about heterogeneity rate across sites as proposed by PartitionFinder (Lanfear et al. Citation2012). We applied two independent runs, each using four Markov chains. Trees were sampled every 100 generations for 10,000,000 generations. After obtaining the convergence, trees from the last 2,447,000 generations were collected to compute the posterior consensus. In the case of TreeFinder (Jobb et al. Citation2004), the separate substitution models were selected for three codon positions according to Propose Model module in this program, and 1000 replicates were assumed in the bootstrap analysis. The posterior probabilities <0.5 and bootstrap percentages <50 were omitted or marked by a dash ‘-’.
The length of Aratinga solstitialis genome (16,984 bp) is identical with Psittacara mitratus. Thectocercus acuticaudatus has the longest of the studied genomes (16,998 bp) and Eupsittula pertinax the shortest (16,980 bp). All mitogenomes revealed the same gene composition, order and orientation. The genomic A + T% of studied Arini is in the range for other avian mitogenomes (Castellana et al. Citation2011). Pyrrhura rupicola genome has the lowest A + T content (52.5%), whereas Aratinga solstitialis has the highest (54.1%). The greatest difference between the maximum and minimum of A + T% was found for tRNA-Tyr gene (7.1%) and the lowest for 12srRNA (0.7%).
The mean percent distance between the genomic sequences is 10.36. The largest difference to others shows Pyrrhura rupicola (about 11%), whereas the closest are pairs Psittacara mitratus – Thectocercus acuticaudatus and Aratinga solstitialis – Ara glaucogularis with 9.7% difference.
Several differences are present in start/stop codons of protein coding genes. Gene nd2 from Eupsittula pertinax uses ATG as start codon, whereas others use ATA. Stop codons of atp8 and nd4L genes from this species end with the truncated TA_ codons instead of the complete TAA in other investigated Arini. The complete TAA stop codon is also present in Psittacara mitratus and Thectocercus acuticaudatus atp6 gene, while in other genera this codon is incomplete (TA_). Only Aratinga solstitialis and Ara glaucogularis nd5 genes end with TAG codon, in contrast to others, which have TAA stop codon.
One extra (not translated) nucleotide, which has been reported for reptile and avian species (Mindell et al. Citation1998; Russell & Beckenbach Citation2008), was also found for all six analysed nd3 genes. However, instead of additional adenine, cytosine was detected in these genes from studied Ara, Aratinga, Psittacara, Pyrrhura, and Thectocercus, whereas thymine in Eupsittula pertinax.
The length of control region (CR) in Aratinga solstitialis (1482 bp) is the same as in Thectocercus acuticaudatus. The shortest CR was found in Psittacara mitratus (1458 bp) and the longest in Pyrrhura rupicola (1492 bp). Small insertion or deletion led to variation in the length of eight intergenic regions (up to 19 bp), as well as two rRNAs (up to 10 bp) and seven tRNAs (up to 4 bp). The tRNA-Val/16S rRNA spacer was identified only in Ara glaucogularis and the atp6/cox3 overlap in Psittacara mitratus and Thectocercus acuticaudatus.
Acknowledgements
We greatly thank MSc Krzysztof Grabowski for technical assistance in the laboratory.
Disclosure statement
The authors report no conflicts of interest and alone are responsible for the content and writing of the paper.
Funding
This work was supported by the National Science Centre Poland under Grant no. [2015/17/B/NZ8/02402].
References
- Castellana S, Vicario S, Saccone C. 2011. Evolutionary patterns of the mitochondrial genome in Metazoa: exploring the role of mutation and selection in mitochondrial protein coding genes. Genome Biol Evol. 3:1067–1079.
- Jobb G, von Haeseler A, Strimmer K. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 4:18.
- Joseph L, Toon A, Schirtzinger EE, Wright TF, Schodde R. 2012. A revised nomenclature and classification for family-group taxa of parrots (Psittaciformes). Zootaxa. 3205:26–40.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Mindell DP, Sorenson MD, Dimcheff DE. 1998. An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles. Mol Biol Evol. 15:1568–1571.
- Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, Escalante AA. 2011. Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol. 28:1927–1942.
- Remsen JV, Schirtzinger EE, Ferraroni A, Silveira LF, Wright TF. 2013. DNA-sequence data require revision of the parrot genus Aratinga (Aves: Psittacidae). Zootaxa. 3641:296–300.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Russell RD, Beckenbach AT. 2008. Recoding of translation in turtle mitochondrial genomes: programmed frameshift mutations and evidence of a modified genetic code. J Mol Evol. 67:682–695.
- Schodde R, Remsen JV, Schirtzinger EE, Joseph L, Wright TF. 2013. Higher classification of New World parrots (Psittaciformes; Arinae), with diagnoses of tribes. Zootaxa. 3691:591–596.
- Urantowka AD. 2016. Complete mitochondrial genome of critically endangered blue-throated Macaw (Ara glaucogularis): its comparison with partial mitogenome of Scarlet Macaw (Ara macao). Mitochondrial DNA. 27:422–424.
- Urantowka AD, Grabowski KA, Strzala T. 2013. Complete mitochondrial genome of Blue-crowned Parakeet (Aratinga acuticaudata)-phylogenetic position of the species among parrots group called Conures. Mitochondrial DNA. 24:336–338.
- Urantowka AD, Mackiewicz P, Strzala T. 2016a. Complete mitochondrial genome of Mitred Conure (Psittacara mitratus): its comparison with mitogenome of Socorro Conure (Psittacara brevipes). Mitochondrial DNA a DNA MappSeq Anal. 27:3363–3364.
- Urantowka AD, Strzala T, Grabowski KA. 2016b. The first complete mitochondrial genome of Pyrrhura sp. – question about conspecificity in the light of hybridization between Pyrrhura molinae and Pyrrhura rupicola species. Mitochondrial DNA. 27:471–473.
