Abstract
Pseudogastromyzon changtingensis belonging to the family Gastromyzontidae is a good model for phylogeny and zoogeography research. In the present paper, the sequenced mitochondrial genome of P. changtingensis is of 16573 bp in length, and encodes 13 typical protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one control region. The result of phylogenetic analysis demonstrates that P. changtingensis is close to that of P. fasciatus.
The Pseudogastromyzon changtingensis belongs to the family Gastromyzontidae, which only lives in China and Southeast Asia. The distinguishing features of this family are pectoral and pelvic fins modified into sucker organs for clinging to objects in fast-flowing streams, and single unbranched anterior ray in pectoral and pelvic fins (Nelson et al. Citation2016). The species P. changtingensis distributes only in Beijiang and Xijiang river systems in China and is a good model for phylogeny and zoogeography research (Yue Citation2000). In this paper, the complete mitochondrial genome of P. changtingensis was determined. The sample of fish was collected from Ganjiang River (26°25′17.9″N, 116°30′44.8″E), Jiangxi province, China. The specimen is stored in the Laboratory of Ichthyology, Shanghai Ocean University, with accession number SLJXWY151023027. The sample DNA is available upon request.
The mitochondrial genome is typically a single circular chromosome in eukaryotes (Zhang et al. Citation2015). In the present study, the sequenced mitochondrial genome of P. changtingensis (GenBank accession number: KX669032) is of 16573 bp in length, and encodes 13 typical protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes (2 tRNAleu, 2 tRNAser), and one control region (CR or D-loop). Most genes are encoded on the H strand, except for the ND6 gene and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAPro, and tRNAGlu), which are encoded on the L strand. The nucleotide composition of mtDNA of P. changtingensis is A (29.45%), T (25.28%), C (28.55%), G (16.73%), which reflects that the percentage of A and T (54.72%) is higher than G and C (45.28%). The lengths of 22 tRNA range from 66 bp (tRNACys) to 76 bp (tRNALys). Apart from COI utilizing GTG, the rest of the 12 protein-coding genes start with the same initiation codon ATG. According to the result given by Mitoannotator (Iwasaki et al. Citation2013), there are seven protein-coding genes with the order of ND2, COII, ATP6, COIII, ND3, ND4, Cytb ending with incomplete stop codon (T-, T-, TA- TA-, T-, TA-, T-), while two genes (COI, ND5) use the stop codon TAA, and ND4L, ND1, ND6 use the stop codon TTA, TAG, TGA, respectively. The end of ATP8 overlaps with the beginning of ATP6 with a length of 10 bp, and there is another overlap between ND5 and ND6 with a length of 4 bp.
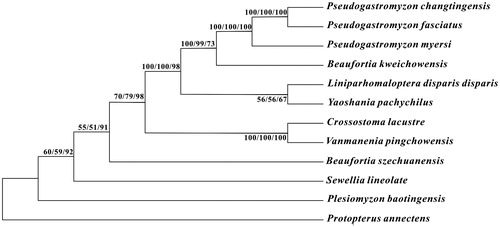
To investigate the phylogenetic relationship among the family Gastromyzontidae, we downloaded the mitochondrial genome sequences of 10 currently available species of Gastromyzontidae, including Beaufortia kweichowensis (KX060617), B. szechuanensis (KP716708), Crossostoma lacustre (AP010774), Liniparhomaloptera disparis disparis (AP013301), Plesiomyzon baotingensis (KF732713), Pseudogastromyzon fasciatus (KX101229), Pseudogastromyzon myersi (AP013300), Sewellia lineolate (AP011292), Vanmanenia pingchowensis (KP005457), and Yaoshania pachychilus (KT031050), together with African lungfish Protopterus annectens (NC018822) as outgroup species. The phylogenetic trees were constructed using MEGA6 (Tamura et al. Citation2013) for neighbour-joining, maximum-likelihood, and maximum parsimony methods. Tree topology was evaluated by 1000 bootstrap replicates. Different methods give the same tree topology, and the result demonstrates that P. changtingensis is close to that of P. fasciatus ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This study was supported by the National Special Basic Research Project of China [No. 2015FY110200] and the National Natural Science Foundation of China [No. 31472280].
References
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Mabuchi K, Takeshima H, Miya M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world. 5th ed. New York (NY): John Wiley and Sons Inc.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yue PQ. 2000. Fauna Sinica, Osteichthyes, Cypriniformes III. Beijing: Science Press.
- Zhang YD, Yu XP, Zhou Z, Huang B. 2015. The complete mitochondrial genome of Acropora aculeus (Cnidarian, Scleractinia, Acroporidae). Mitochondrial DNA Part A. [Epub ahead of print]. DOI: 10.3109/19401736.2015.1082092.