Abstract
Androglossini is one of the four tribes recognized within the group of neotropical parrots. The tribe includes 10 genera of which Amazona genus is presently represented by about 30 species. The number of species may increase in the future because paraphyly of some Amazona species was recently demonstrated and some subspecies were proposed to be elevated to the species rank. Evolutionary history of Amazona genus also remains unresolved because published phylogenies suggest contradictory scenarios concerning directions of islands-mainland colonization. Therefore, we sequenced mitogenome of Amazona ventralis from Greater Antilles to gain molecular data essential in future examination of this genus diversification.
Neotropical parrots, i.e. Arinae subfamily are divided into four tribes (Schodde et al. Citation2013). Tribe Androglossini, which grouped 10 genera including Amazona represented by the largest number of species distributed in Central and South America, and the Caribbean. Strong-heavy bill, short-rounded tail, prominent naked cere, and a distinct notch in the upper mandible are characteristic features for this genus. Except for Lesser Antillean (arausiaca, guildingii, imperialis, versicolor) and one continental (vinacea) species, their body plumage is predominantly green with variable colorations on the head, breast, wing coverts, and/or flight feathers.
Based on the variation of theses accenting colours, 27–29 Amazona species were primarily recognized depending on different treatments of Yellow-Headed Amazon (YHA) group, as a single (ochrocephala) or three species (ochrocephala, auropalliata, oratrix). Resurrection of the new Alipiopsitta genus from Amazona xanthops decreased the species number, which increased again with the elevation of Amazona rhodocorytha and Amazona kawalli from subspecies to species status based on genetics (Rusello & Amato Citation2004). Amazona tresmariae was distinguished from A. oratrix also based on results of molecular analyses (Eberhard & Bermingham Citation2004). The number of species may further increase because paraphyly of auropalliata, ochrocephala and oratrix species was recently revealed (Urantówka et al. Citation2014). Moreover, even A. ochrocephala ochrocephala occurred to be paraphyletic with respect to A. aestiva and A. barbadensis. Additionally, Wenner et al. (Citation2012) suggested that two Amazona farinosa subspecies (virenticeps and guatemalae) should be elevated to species rank.
Evolutionary history of Amazona genus also remains unresolved because some clades in current phylogenies occurred biogeographically heterogeneous. Central American taxa group with South American ones, whereas Antillean species are clustered with continental taxa (Rusello & Amato Citation2004; Ottens-Wainright et al. Citation2004). So far, a clear evolutionary scenario was proposed only for continental YHA taxa and Lesser Antillean species (arausiaca and versicolor). It postulates that island parrots colonized the mainland. However, the scenario for Greater Antillean species is still controversial. Phylogenies based on CYTB gene placed Central American Amazona albifrons basal to all Greater Antillean species suggesting colonization of islands from mainland (Ottens-Wainright et al. Citation2004). However, other phylogenies based on mitochondrial and nuclear markers placed Jamaican species (agilis) basal to the clade with mainland and other Greater Antillean species (Rusello & Amato Citation2004; see also ) suggesting more complex colonizations. This indicates that more molecular data are required to reconstruct the precise Amazona phylogeny. So far, complete mitogenomes of only two Amazona species (barbadensis and ochrocephala) are available (Urantowka et al. Citation2013; Eberhard & Wright Citation2016). Therefore, we sequenced Greater Antillean Amazona ventralis mitogenome with 18,571 bp (GeneBank accession number KX925977) to gain appropriate molecular data for examination of Amazona genus diversification.
Figure 1. The phylogenetic tree obtained in MrBayes for cytb gene indicating that the studied individual (bolded) belongs to Amazona ventralis. The blood sample from which DNA was isolated is available in the laboratory at the Department of Genetics in Wroclaw University of Environmental and Life Sciences under the number ADUAKPM01. Clades with taxa inhabited different geographic regions were marked by various shading/colours: CA: Central America; GA: Greater Antilles; LE: Lesser Antilles; SA: South America; TM: Tres Marías Islands. An internal placement of taxa from Lesser Antilles and Central America within South American parrots implies that South America mainland was the source of those colonizations. Similarly, Tres Marías Islands were invaded from Central America, whereas some Central American parrots (e.g. A. albifrons) can have also Greater Antillean origin. Values at nodes, in the order shown, indicate posterior probabilities found in MrBayes (PP) and bootstrap percentages calculated in TreeFinder (BP). In the MrBayes (Ronquist et al. Citation2012) analysis, separate mixed substitution models were assumed for three codon positions with information about heterogeneity rate across sites as proposed by PartitionFinder (Lanfear et al. Citation2012). We applied two independent runs, each using four Markov chains. Trees were sampled every 100 generations for 10,000,000 generations. After obtaining the convergence, trees from the last 3,527,000 generations were collected to compute the posterior consensus. In the case of TreeFinder (Jobb et al. Citation2004), the separate substitution models were selected for three codon positions according to Propose Model module in this program, and 1000 replicates were assumed in the bootstrap analysis. The posterior probabilities <0.5 and bootstrap percentages <50 were omitted or marked by a dash ‘-’.
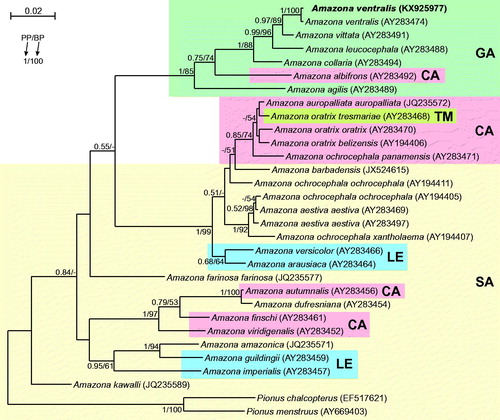
Although morphology of the analyzed specimen (Polish captive bird) was absolutely typical for ventralis individuals, to prove its species belonging, we compared its cytochrome b sequence with those from other Amazona taxa. The obtained tree () revealed that the analyzed individual grouped significantly with another representative of its species and this clade was sister to Puerto Rican Amazona vittata.
Disclosure statement
The authors report no conflicts of interest and alone are responsible for the content and writing of the paper.
Funding
This work was supported by the National Science Centre Poland (Narodowe Centrum Nauki, Polska) under Grant no. [2015/17/B/NZ8/02402].
References
- Eberhard JR, Bermingham E. 2004. Phylogeny and biogeography of the Amazona ochrocephala (Aves: Psittacidae) complex. Auk. 121:318–332.
- Eberhard JR, Wright TF. 2016. Rearrangement and evolution of mitochondrial genomes in parrots. Mol Phylogenet Evol. 94:34–46.
- Jobb G, von Haeseler A, Strimmer K. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 4:18.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Ottens-Wainright P, Kenneth MH, Eberhard JR, Burke RI, Wiley JW, et al. 2004. Independent geographic origin of the genus Amazona in the West Indies. J Caribbean Ornithol. 17:23–49.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Russello MA, Amato G. 2004. A molecular phylogeny of Amazona: implications for Neotropical parrot biogeography, taxonomy, and conservation. Mol Phylogenet Evol. 30:421–437.
- Schodde R, Remsen JV, Schirtzinger EE, Joseph L, Wright TF. 2013. Higher classification of New World parrots (Psittaciformes; Arinae), with diagnoses of tribes. Zootaxa. 3691:591–596.
- Urantowka AD, Hajduk K, Kosowska B. 2013. Complete mitochondrial genome of endangered Yellow-shouldered Amazon (Amazona barbadensis): two control region copies in parrot species of the Amazona genus. Mitochondrial DNA. 24:411–413.
- Urantówka AD, Mackiewicz P, Strzała T. 2014. Phylogeny of Amazona barbadensis and the Yellow-headed Amazon complex (Aves: Psittacidae): a new look at South American parrot evolution. PLoS One. 9:e97228.
- Wenner TJ, Russello MA, Wright TF. 2012. Cryptic species in a Neotropical parrot: genetic variation within the Amazona farinosa species complex and its conservation implications. Conservation Genetics. 13:1427–1432.
