Abstract
The complete mitochondrial genome of Diadema setosum was sequenced by using long-PCR and primer walking methods. The complete mitochondrial genome is a 15,708 bp circular molecule, which contains 22 tRNAs, 2 rRNAs, and 13 protein-coding genes as well as a non-coding control region as in other urchin. The genome was composed of 27.5% A, 30.7% T, 24.3% C, and 17.5% G. The control region is 133 bp. Twelve of the 13 mitochondrial protein-coding genes use ATG as their start codon while the ATP8 gene starts with GTG. Six of the 13 mitochondrial protein-coding genes use TAA as their complete stop codon while seven of them use TAG as their complete stop codon.
Diadema setosum belongs to class Echinoidea, order Aulodonta, family Diadematidae, genus Diadema. In China, it is mainly distributed in Hainan, Xisha Islands and other places; its worldwide distribution is mainly in the South Pacific and the Indian Ocean, from Africa, Madagascar, South China Sea to Japan and other places (Zhang et al. Citation1957; Addison & Hart Citation2002; Liao & Clark Citation1995). Most of the studies are focused on its morphology and ecological resources (You et al. Citation2004; Liao & Xiao Citation2011; Liu. Citation2014; Zhang. Citation2011). No research on its complete mitochondrial genome has been documented till now (Zeng et al. Citation2012). Therefore, it is highly important for the further studies.
In this study, we sequenced the entire mitochondrial genome of D.setosum for the first time (GenBank accession number: KX385835) and shares high similarity on gene content and structure with most sea urchin. So far, mitochondrial genomes have been sequenced in 19 sea urchin species, but none in Aulodonta. So the addition of the mitogenome of D.setosum will enrich the genomic information of the Aulodonta.
The sample was obtained from XiSha Islands offshore (16°32 'north latitude and longitude 111°40') and total DNA was extracted from the Muscle tissue between the teeth. The sample currently stored in the Fisheries Biology Research Laboratory, Ocean University of China, the number is YWQD-QFD-20140514-X-010. The mitochondrial DNA were amplified through long-PCR and primer walking methods with primer sets designed from the conserved regions of the known mitogenome sequences of sea urchin (Fu et al. Citation2014; Chen et al. Citation2012; Jung et al. Citation2013; Li et al. Citation2013). The PCR products were sequenced, and assembled and annotated using DNASTAR Lasergene v7.1.0.
The mitogenome of D.setosum is a circular molecule of about 15,708 bp in length, consisting of 22 tRNA genes, 2 rRNA genes (12S rRNA and 16S rRNA), and 13 typical vertebrate protein-coding regions generally found in other echinoderm species. This study showed that all mitochondrial genes were encoded on the H-strand with the exception of five tRNA genes (tRNAGln, tRNAAla, tRNAVal, tRNAAsp, tRNASer (UCN)) and one protein-coding gene ND6, which were encoded on the L-strand. The overall nucleotide composition of the heavy strand was as follows: 27.5% A, 30.7% T, 24.3% C, and 17.5% G, with anti-G bias. According to the two formulas, AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) (Liu et al. Citation2007), the AT-skew of D.setosum was −0.0565, and its GC-skew was −0.1624. The 13 protein-coding genes are totally 11202 bp in size, accounting for 71.33% of the whole mitogenome. The two rRNAs, 12S rRNA, and 16S rRNA, were 895 and 1,529 bp in length, respectively. 22 tRNAs of D.setosum ranging from 67 to 73 bp, interspersed between the rRNA and protein-coding genes, similar to other mitochondrial genomes in sea urchins. Twelve of the 13 mitochondrial protein-coding genes use ATG as their start codon while the ATP8 gene starts with GTG. Six of the 13 mitochondrial protein-coding genes use TAA as their complete stop codon while seven of them use TAG as their complete stop codon.
Ten kinds of echinoidea sea urchin complete mitochondrial genome sequences (not including ND6 gene, the termination of protein-coding genes, and non-coding regions) after splicing sequence length of about 15000 bp in sequence (). To outline of rough sea cucumber apostichopus (Stichopus horrens) as the out group species, build echinoidea mitochondrial genome sequence of all ML phylogenetic tree (). Based on the complete mitochondrial genome of 10 sea urchins, it established the maximum likelihood(ML) of phylogenetic tree. The result shows that the genetic relationship between Diadema setosum and Echinothrix diadema is mostly related. The sea urchins of Aulodonta is closer to Spatangoida than Camarodonta. This study confirms the phylogenetic relatives of sea urchins.
Figure 1. ML phylogenetic tree established based on complete mitogenome sequences. Build the ML of phylogenetic tree branches since the fair value is very high, most of 100, to build the ML phylogenetic tree credibility is higher.
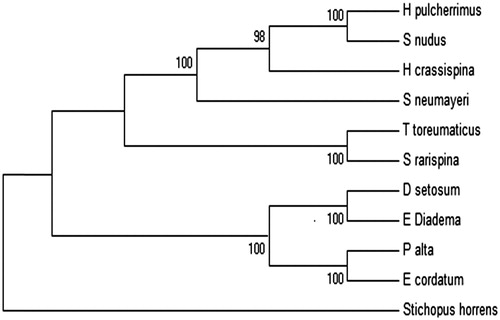
Table 1. The complete mitochondrial genome of Echinoidea.
Acknowledgements
We are grateful to Dr Shichun Sun for the sample collection.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Addison JA, Hart MW. 2002. Characterization of microsatellite loci in sea urchins (strongylocentrotus spp.). Mol Ecol Notes. 2:493–494.
- Chen J, Ma GQ, Miao ZQ, Shui BM, Gao TX. 2012. Complete mitochondrial genome sequence of the spinyhead croaker Collichys lucidus (Perciformes, Sciaenidae) with phylogenetic consideration. Mol Biol Rep. 39:4249–4259.
- Fu W, Liu C, Wu G, Zeng X. 2014. The complete mitochondrial genome of Temnopleurus hardwickii (Camarodonta: Temnopleuridae). Mitochondrial DNA. 1–2.
- Jung GL, Ghoi HJ, Pae SJ. 2013. Complete mitochondrial genome sequence of the sea urchin: Mesocentrotus nudus (Strongylocentrotidae: Echinoida). Mitochondrial DNA. 24:466–468.
- Li Y, Pan XZ, Song N, Yanagimoto T, Gao TX. 2013. The Complete mitochondrial genome of Japanese Konosirus punctatus (Clupeiformes: Clupeidae). Mitochondrial DNA. 24:481–483.
- Liao YL, Clark AM. 1995. The echinoderms of Southern China. Beijing: Science Press; p. 347–351.
- Liao YL, Xiao N. 2011. Species compositon and faunal characteristics of echinoderms in China seas. Biodiversity Science. 19:729–736.
- Liu CL. 2014. The study on mitochondrial genome and phylogenetic analysis of Temnopleuridae. Qindao: Ocean University of China.
- Liu XH, Huang JQ, Zhou ZC, Dong Y. 2007. Sequence analysis of mtDNA 16S rRNA gene fragments among five species of sea urchins. Fish Sci. 26:331–334.
- You XL, Liao YL, Sun S. 2004. The sea urchins from the continental shelf of the East China Sea. Studia Marina Sinica. 46:181–201.
- Zeng XQ, Zhang WF, Gao TX. 2012. Molecular phylogenetics of genus Temnopleurus based on 16S rRNA and COL sequences. Period Ocean Univ China. 42:47–51.
- Zhang FY, Wu BL, Liao YL. 1957. China’s sea urchin. J Biol. 7:18–25.
- Zhang WF. 2011. Studies on the morphology and genetics of Temnopleurus and the taxonomy of Temnopleuridae inshore water of China Sea. Qingdao: Ocean University of China.
