Abstract
The complete chloroplast genome sequence of an important medicinal plant of the family Ranunculaceae, Aconitum carmichaelii Debx., was characterized in this study. The assembled chloroplast genome was 154,776 bp in length, which included a large single-copy (LSC), a small single-copy (SSC), and two inverted repeat (IR) regions of 86,330bp, 15,986 bp, and 26,193 bp, respectively. The GC content of the genome was 38.1%. Phylogenetic analysis with the whole nucleotide sequences of reported Aconitum chloroplast genomes indicated a close relationship of A. carmichaelii with A. kusnezoffii.
Aconitum carmichaelii Debx. belongs to genus Aconitum of family Ranunculaceae. Aconitum species, which contain various terpenoids, alkaloids, phenypropanoids, and polysaccharides, are well-known in East Asia for their toxicity and as potent herbal medicines (Kadota Citation1987). Its lateral roots, named as Fuzi, have been extensively used as cardiotonic, analgesic, anti-inflammatory, and diuretic agents to treat colds, polyarthralgia, diarrhoea, heart failure, beriberi, and oedema (Murayama et al. Citation1991). While the main roots, named as Chuanwu, are used for the clinical treatment of pains and rheumatics (Liu et al. Citation2012). Due to the similar morphology and drug confusion among Aconitum plants, it is very important to carry out identification and phylogeny studies of A. carmichaelii. Nineteen Aconitum species have been identified using the chloroplast genome intergenic region psb-trnH (He et al. Citation2010). The phylogenetic relationship of 31 genera of Ranunculaceae have been analyzed applying chloroplast DNA restriction site variation (Johansson Citation1995). Whereas, more studies on the chloroplast genome are needed for the complete molecular identification. In this study, we characterized the complete chloroplast genome sequence of A. carmichaelii to contribute to further molecular and phylogenetical studies of this plant species.
Aconitum carmichaelii was collected from Jiangyou GAP (Good Agricultural Practice of Medicinal Plants and Animals) plantation bases of Sichuan province and deposited in The State Bank of Chinese Drug Germplam Resources. The complete chloroplast genome of A. carmichaelii was 154,776 bp in size (GenBank accession number KY006977) with 38.1% overall GC content. The genome structure was highly similar to the reported chloroplast genome of closely related species A. chiisanense (Lim et al. Citation2015), and contained 83 protein-coding genes, 35 tRNA genes, and 8 rRNA genes. The chloroplast genome was composed of a large single-copy (LSC), a small single-copy (SSC), and two inverted repeat (IR) regions of 86,330bp, 15,986 bp and 26,193 bp, respectively. Thirty-three genes (14 tRNA, 8 rRNA, and 11 protein-coding genes) were duplicated in two inverted repeat regions. As in the chloroplast genomes of other Ranunculaceae species reported (Lim et al. Citation2015; Park et al. Citation2015), the rpl32 gene was lost in A. carmichaelii.
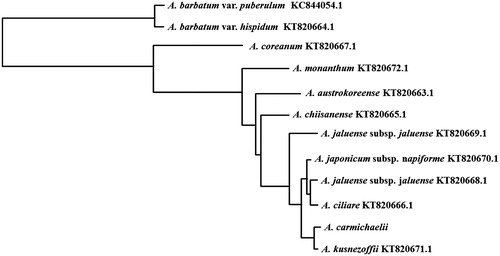
A phylogenetic tree of A. carmichaelii was constructed based on complete chloroplast genomes of 12 Aconitum species by a neighbour-joining analysis using the BLAST pairwise alignments and Blast Tree View tool in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) (). The phylogenetic tree indicated that A. carmichaelii has a closer genetic relationship with A. kusnezoffii, which belongs to the series Inflata Steinb., section Aconitum, subgenus Aconitum, as well as our target plant.
Disclosure statement
The authors declare that there is no conflict of interest in the preparation and execution of this manuscript.
Additional information
Funding
References
- He J, Wong KL, Shaw PC, Wang H, Li DZ. 2010. Identification of the medicinal plants in Aconitum L. by DNA barcoding technique. Planta Med. 76:1622–1628.
- Johansson JT. 1995. A revised chloroplast DNA phylogeny of the Ranunculaceae, in systematics and evolution of the Ranunculiflorae. Springer. 9:253–261.
- Kadota Y. 1987. A revision of Aconitum subgenus Aconitum (Ranunculaceae) of East Asia. Japan: Sanwa Shoyaku Company.
- Lim CE, Kim GB, Baek S, Han SM, Yu HJ, Mun JH. 2015. The complete chloroplast genome of Aconitum chiisanenese Nakai (Ranunculaceae). Mitochondrial DNA. 1–2.
- Liu XX, Jian XX, Cai XF, Chao RB, Chen QH, Chen DL, Wang XL, Wang FP. 2012. Cardioactive C19-diterpenoid alkaloids from the lateral roots of Aconitum carmichaeli “Fu Zi”. Chem Pharm Bull (Tokyo). 60:144–149.
- Murayama M, Mori T, Bando H, Amiya T. 1991. Studies on the constituents of Aconitum species. IX. The pharmacological properties of pyro-type aconitine alkaloids, components of processed aconite powder ‘kako-bushi-matsu’: analgesic, antiinflammatory and acute toxic activities. J Ethnopharmacol. 35:159–164.
- Park S, Jansen R, Park S. 2015. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 15:40.