Abstract
The complete mitogenome sequence of Microphysogobio amurensis has been amplified and sequenced in this study. The overall base composition of M. amurensis mitogenome is: A (30%), C (26.8%), G (17.2%), T (26%), with an obvious A + T bias (56%). The assembled mitogenome, consisting of 16605 bp, has 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. To estimate the status of M. amurensis, all available mitogenomes of Microphysogobio were downloaded from GenBank for phylogenetic analysis. The result strongly supported the traditional taxonomic hypothesis that M. amurensis was sister to M. brevirostris and M. koreensis. However, taxonomic confusion on its generic assignment remains to be further studied, as this study and other relevant molecular studies not only suggested that Microphysogobio could be divided in two major clusters (viz. the Palearctic species group and the Oriental species group), but also that two allied genera (Biwia, Rostrogobio) were embedded within the Palearctic species group of Microphysogobio.
The genus Microphysogobio Mori Citation1934, generally including a number of diminutive, rheophilic, and benthic cyprinid taxa referred to as dwarf gudgeon, represents as a geographically widespread but poorly diagnosed group and presents many challenges to authors, who hope to address the species level phylogeny of this genus and allies (Tang et al. Citation2011; Huang et al. Citation2016). The northerly outrider of the genus, M. amurensis (Taranetz Citation1937) was initially described as a new genus and species Rostrogobio amurensis (type location: the middle and lower Amur River, Khanka Lake), but some later authors (e.g. Reshetnikov Citation2003, Bogutskaya & Naseka Citation2004) considered R. amurensis a junior synonym of M. tungtingensis (Nichols Citation1926), which was described from Tungting Lake, a tributary of Yangtze River, southern China. In contrast to this opinion, Bǎnǎrescu & Nalbant Citation1966, Citation1973) gave it a subspecific designation of M. tungtingensis and extended its distribution to northeastern China and Russia, excluding the Korean Peninsula inhabited by another two tungtingensis-like Microphysogobio. Recently, Kottelat (Citation2006) questioned the synonymy of M. amurensis under M. tungtingensis, and the differentiation was subsequently confirmed by evidence based on the partial region of mitochondrial DNA (Tang et al. Citation2011). Although these molecular and morphological analyses have revisited the phylogenetics of the genus Microphysogobio and advanced our understanding of intrageneric evolution, most are still inconclusively resolved, including the reliable phylogenetic relationships of M. amurensis. In this study, therefore, we determined the complete mitogenome of M. amurensis, the first representative of the nominal genus Rostrogobio, and analyzed its phylogenetic relationships within subfamily Gobioninae.
DNA was extracted from M. amurensis (topotypic specimen SCAU 1179906) collected from the Khanka (Xingkai) Lake, Amur River basin (45°11′47″N, 132°14′32″E) and deposited in the collection of South China Agricultural University (SCAU). The complete mitogenome sequence was amplified using long-range PCR and sequenced using the primer-walking strategy.
The complete mitochondrial genome of M. amurensis was 16,605 bp in size (GenBank accession number KY228977), which consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region. The overall base composition of the entire genome was 30% for A, 26.8% for C, 17.2% for G, and 26% for T and had low G + C content of 44%. All PCGs started with typical ATG codons, except for the COX1 gene, which started with GTG. Six of the 13 PCGs harboured the truncated termination codon T–– or TA–, three genes stop with TAA (COX1, ATP6, ND4L), four with TAG (ND1, ND2, ATP8, ND5). The longest one was ND5 gene (1836 bp) in all PCGs, whereas the shortest was ATP8 gene (165 bp). The length of 12S rRNA and 16S rRNA genes were 958 bp and 1689 bp, respectively. The control region was 929 bp in size and located between tRNA-Pro and tRNA-Phe.
To estimate the status of M. amurensis, all available mitogenomes of Microphysogobio were downloaded from GenBank for phylogenetic analysis. Our molecular phylogeny shows that the putative members of Microphysogobio from East Asia appear in two geographically distinct clades with high statistical support, namely the Palearctic clade and the Oriental clade (). The Palearctic clade includes the dwarf gudgeons that are widely distributed in Russian Far East, Mongolia, the Korean Peninsula and the nearby mainland of northern China, as well as northern Taiwan Island; while the Oriental clade joins the taxa of southern China (including central and southern Taiwan Island). Strikingly, the species belonging to these two clades have overlapping ranges in the Yangtze River basin and paleo-Min River basin, which comprise the transitional zone between the Palearctic realm and the Oriental realm (Mori Citation1936; Bǎnǎrescu Citation1992). The overall genetic architecture further showed that putative members of Microphysogobio occurring in northern China and adjacent area (e.g. M. amurensis) are more closely related to Japanese endemic Biwia zezera (type species of Biwia Jordan & Fowler Citation1903) than to their southern Chinese cousins (e.g. M. tafangensis), indicating that both Microphysogobio and Biwia are taxonomically heterogeneous. This finding is consistent with several phylogenetic results based on mitochondrial and nuclear DNA sequences (Kim & Bang Citation2010; Tang et al. Citation2011). Further proof of this are the degree of divergences between each species of Microphysogobio ranging from 7% to 15% in the whole mitogenomes of uncorrected pairwise genetic distances, which greatly overlapped the range (9%–14%) observed between B. zezera and all known sibling species of Microphysogobio. As such, the species currently classified under the genus Microphysogobio are clearly in a need of taxonomical revision.
Figure 1. Phylogenetic relationships among M. amurensis and other Asian dwarf gudgeons with complete mitogenome sequences on GenBank were inferred using the Maximum-Likelihood (ML), Neighbour-Joining (NJ), and Bayesian (BA) phylogenetic analyses, respectively. Numbers above branches are bootstrap support of the ML, NJ analysis higher than 95 and posterior probabilities of the Bayesian analysis higher than 95%, for the combined analysis. The results for the entire mitogenomes gave with three methods very similar tree topologies. The gene’s accession numbers for tree construction are listed as follows: M. tafangensis (KF857260.1), M. alticorpus (KC762939.1), M. fukiensis (KJ933414.1), M. brevirostris (KF319122.1), M. yaluensis (KR075133.1), M. koreensis (JX179157.1), M. longidorsalis (AP011394.1), Biwia zezera (AB250107.1), B. yodoensis (AB250108.1), B. springeri (AP011360.1), Abbottina rivularis (AP011257.1), Saurogobio dabryi (KF534790.1), Pseudogobio esocinus (AP009310.1). Clades with taxa inhabited different biogeographic realms were marked by various shading/colours (Oriental: the Oriental species; Palearctic: the Palearctic species). The internal placements of Biwia taxa from Japan and Korea within the genus Microphysogobio implied that their independent generic positions were not supported herein, and they probably could be placed in a single genus. If the priority status of Microphysogobio vs. Biwia is confirmed, the latter being a senior synonym.
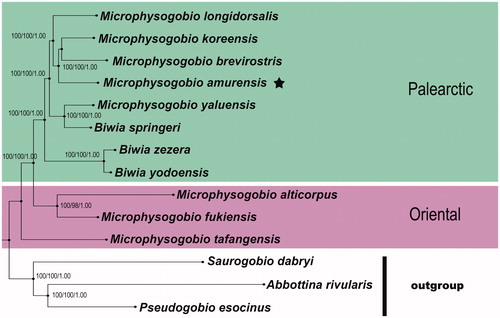
Unfortunately, the issue that profoundly influences taxonomic limbo and confounds the systematic of Microphysogobio and allied genera to date is loss of the type specimen of M. hsinglungshanensis (type species of Microphysogobio); even none of the genetic data of M. hsinglungshanensis is virtually available for corroborating the validity of Microphysogobio as a distinct genus (Jiang et al. Citation2012; Jiang & Zhang Citation2013). Recent molecular studies of the Asian gudgeons, however, have examined many of the Microphysogobio taxa that have been considered closely related to M. hsinglungshanensis (including M. chinssuensis, which is regarded as a synonym or counterpart of M. hsinglungshanensis by the majority of authors). Their results were in accordance with the aforementioned phylogenetic relationships (Yang et al. Citation2006; Liu et al. Citation2010; Tang et al. Citation2011). According to its original description by Mori (Citation1934), M. hsinglungshanensis was known from Hsinglung County, bordering Beijing, northern China that belongs to the Palearctic region in biogeography, hence we could not eliminate the possibility that this name-bearing type species may fall into the Palearctic clade with B. zezera and M. amurensis. Taken together, it seems probable that the nominal genus Biwia consists of the autapomorphic species of Microphysogobio (including Rostrogobio) and does not warrant separate status.
Here, we choose to recognize ‘Microphysogobio’ amurensis with this genus in single quotes until the complete taxonomic revision of Biwia-Microphysogobio complex is elucidated, but we will not be surprised if this species is reallocated into the genus Biwia in the future.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
References
- Bǎnǎrescu PM. 1992. A critical updated checklist of Gobioninae (Pisces, Cyprinidae). Travaux Du Museum D?histoire Naturelle “Grigore Antipa”. 32:303–330.
- Bǎnǎrescu PM, Nalbant TT. 1966. Revision of the genus Microphysogobio (Pisces, Cyprinidae). Véstnik Československé společnosti zoologické. 30:194–209.
- Bǎnǎrescu PM, Nalbant TT. 1973. Pisces, Teleostei. Cyprinidae (Gobioninae). Das Tierreich. 93:1–304.
- Bogutskaya NG, Naseka AM. 2004. Catalogue of agnathans and fishes of fresh and brackish waters of Russia with comments on nomenclature and taxonomy. Moscow: KMK Scientific Press, 389 pp. [in Russian]
- Huang SP, Chen IS, Shao KT. 2016. A new species of Microphysogobio (Cypriniformes: Cyprinidae) from Fujian Province, China, and a molecular phylogenetic analysis of Microphysogobio species from southeastern China and Taiwan. Proc Biol Soc Wash. 129:195–211.
- Jiang ZG, Gao EH, Zhang E. 2012. Microphysogobio nudiventris, a new gudgeon species of cyprinid fishes (Teleostei: Cypriniformes) from Hubei Province, South China. Zootaxa. 3586:211–221.
- Jiang ZG, Zhang E. 2013. Molecular evidence for taxonomic status of the gudgeon genus Huigobio Fang, 1938 (Teleostei: Cypriniformes), with a description of a new species from Guangdong Province, South China. Zootaxa. 3731:171–182.
- Jordan DS, Fowler HW. 1903. A review of the cyprinoid fishes of Japan. Proc U S Nat Mus. 26:811–862.
- Kim KY, Bang IC. 2010. Molecular phylogenetic position of Abbottina springeri (Cypriniformes; Cyprinidae) based on nucleotide sequences of RAG1 gene. Korean J Ichthyol. 22:273–278.
- Kottelat M. 2006. Fishes of Mongolia, a check-list of the fishes known to occur in Mongolia with comments on systematics and nomenclature. Washington (DC): World Bank Report (NEMO), i–xi, p. 103.
- Liu HZ, Yang JQ, Tang QY. 2010. Estimated evolutionary tempo of East Asian gobionid fishes (Teleostei: Cyprinidae) from mitochondrial DNA sequence data. Chin Sci Bull. 55:1501–1510.
- Mori T. 1934. The fresh water fishes of Jehol. In: Tokunaga S, editor. Report of the first scientific expedition to Manchoukuo. Vol. 1. Tokyo: Office of the Scientific expedition to Manchoukuo; p. 1–61.
- Mori T. 1936. Studies on the geographical distribution of freshwater fishes in Eastern Asia. Chosen: Keijo Imperial University.
- Nichols JT. 1926. Some Chinese fresh-water fishes. XV. Two apparently undescribed catfishes from Fukien. XVI. Concerning gudgeons related to Pseudogobio, and two new species of it. XVII. Two new Rhodeins. Am Mus Novit. 214:1–7.
- Reshetnikov YS. 2003. Atlas of Russian freshwater fishes, vol. 1. Moscow: Nauka, p. 379. [in Russian]
- Tang KL, Mary KA, Chen WJ, Hirt MV, Morgan ER, Tetsuya S, Leah MS, Yang L, Henry LB, He SP, et al. 2011. Phylogeny of the gudgeons (Teleostei: Cyprinidae: Gobioninae). Mol Phylogenet Evol. 61:103–124.
- Taranetz AY. 1937. A note on a new genus of gudgeons from the Amur Basin. Izvestiia Tikhookeanskogo nauchnogo institute rybnogo khoziaistva [Bulletin of the Far Eastern Branch of the Academy of Sciences of the USSR]. 23: 113–115.
- Yang J, He S, Freyhof J, Witte KE, Liu H. 2006. The phylogenetic relationships of the Gobioninae (Teleostei: Cyprinidae) inferred from mitochondrial cytochrome b gene sequences. Hydrobiologia. 553:255–266.
