Abstract
Dryophytes suweonensis is an endangered species with fragmented and declining populations from the Korean peninsula. We described 17,448 bp of D. suweonensis mtDNA, which had a shorter D-loop than other closely related species. The variation in nucleotide composition was similar to that of Hyla tsinlingensis but was larger than the one of its sister clade, D. japonicus.
Keywords:
Comparative genetic analyses between the widespread Dryophytes japonicus and the Endangered D. suweonensis are scarce, only specifying that the two species diverged about 5 mya (Li et al. Citation2015). Variations in behavioural ecology are now better understood, with clear behavioural and ecological differentiations between the two species (Jang et al. Citation2011; Borzée et al. Citation2016). Sequencing the full mtDNA loop of D. suweonensis will enable the future development of primers to help identify populations of importance for conservation.
One D. suweonensis was caught in Pyeongtaek, Republic of Korea (37.001°N; 127.0055°E) in June 2015 for buccal swabbing. DNA was extracted (Qiagen DNeasy, Hilden, Germany) following the instructions of the manufacturer. Primers for two long and accurate (LA) PCRs were designed based on Hyla tsinlingensis mtDNA (GenBank accession KP212702), with ad hoc primers for the D-loop due to size difference with other hylids (Geneious v. R 9; Auckland, New Zealand). Sanger sequencing was conducted by Cosmogenetech (Seoul, Republic of Korea), before GenBank upload under accession number KX854020.
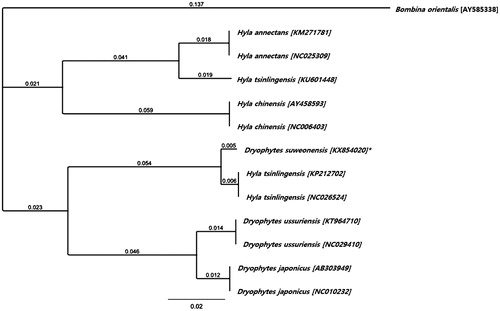
The nucleotide composition for D. suweonensis was 29.3% of A, 27.2% of C, 15.0% of G, and 28.6% of T. The total nucleotide composition was 17,448 bp long, thus 847 bp shorter than Hyla tsinlingensis, with only 0.1% variation in the C nucleotide frequency (Huang et al. Citation2014). However, when compared with D. japonicus (accession number IABHU6123), it was 2071 bp shorter, with a variation in nucleotide frequency of 0.3% of A, 1.8% of C, 0.5% of G, and 1.9% of T. Using the Geneious Tree Builder plugin on Geneious v. R 9, a Neighbour-Joining Tree was constructed under the Tamura–Nei model and a 65% (5/−4) cost matrix, based on the complete mitogenetic sequences of all 11 Hyla uploaded on GenBank (), and with Bombina orientalis as an outgroup. The grouping of D. suweonensis and two H. tsinlingensis together, while another H. tsinlingensis is clustered within the Hyla clade, indicates a potential miss-identification of the first two H. tsinlingensis individuals. An alternative identification would be D. immaculatus, due to the close relatedness with D. suweonensis (Li et al. Citation2015), and thus highlighting the divergence between these two species.
Disclosure statement
None of the co-authors has any conflict of interest to declare. The experiments in this study comply with the current laws of the Republic of Korea (Ministry of Environment Permit Number 2015-28), and did not allow for voucher individual collection due to ethical reasons.
Additional information
Funding
References
- Li J-T, Wang J-S, Nian H-H, Litvinchuk SN, Wang J, Li Y, Rao D-Q, Klaus S. 2015. Amphibians crossing the bering land bridge: evidence from holarctic treefrogs (Hyla, Hylidae, Anura). Mol Phylogenet Evol. 87:80–90.
- Jang Y, Hahm EH, Lee H-J, Park S, Won Y-J, Choe JC. 2011. Geographic variation in advertisement calls in a tree frog species: gene flow and selection hypotheses. PLoS One. 6:e23297.
- Borzée A, Kim JY, Jang Y. 2016. Asymmetric competition over calling sites in two closely related treefrog species. Sci Rep. 6:32569.
- Huang M, Duan R, Tang T, Zhu C, Wang Y. 2014. The complete mitochondrial genome of Hyla tsinlingensis (Anura: Hylidae). Mitochondrial DNA. 27:4130–4131.