Abstract
In this study, we analyzed the complete mitochondrial genome of the cavity-nesting honeybee, A. koschevnikovi. The mitochondrial genome of A. koschevnikovi was observed to be a circular molecule of 15,278 bp and was similar to that of the other cavity-nesting honeybee species. The average AT content in the A. koschevnikovi mitochondrial genome was 84%. It was predicted to contain 13 protein-coding, 24 tRNA and two rRNA genes, along with one A + T-rich control region, besides three tRNA-Met repeats.
The Southeast Asian honeybee, Apis koschevnikovi, belonging to subgenus Apis is a cavity-nesting honeybee of Asia belonging to a group that includes A. cerana, A. nigrocincta, and A. nuluensis along with the European-African A. mellifera. Molecular phylogenetic analyses of the partial DNA sequences indicate that this species and A. cerana are sister taxa (Arias et al. Citation1996; Tanaka et al. Citation2001). The distribution of this species has been geographically restricted, mainly in the isolated tropical evergreen straits of Borneo, Malay Peninsula and Sumatra (Hadisoesilo et al. Citation2008). Basic knowledge on this species has been lacking; little information is available on the genetic diversity and population density, which is necessary for conservation of the species (Tanaka et al. Citation2003).
Adult worker was collected from a hive in an apiary at the Agriculture Research Station Tenom in Sabah, Malaysia (the specimen is stored in the National Museum of Nature and Science, Japan accession number: NSMT-I-HYM742391). Genomic DNA isolated from one worker was sequenced using Illumina’s Next Seq 500 (Illumina). The resultant reads were assembled and annotated using the MITOS web server (Bernt et al. Citation2013, Germany), the MEGA6 software (Tamura et al. Citation2013) and GNETYX v.10 (Genetyx Corporation, Japan). The phylogenetic analysis was performed using the TREEFINDER v.2011 (Jobb et al. Citation2004) based on the nucleotide sequences of 13 protein-coding genes.
The A. koschevnikovi mitochondrial genome forms a closed loop that is 15,278 bp long (accession number AP017643). The A. koschevnikovi mitochondrial genome represents a typical hymenopteran mitochondrial genome and matches the common organization present in the bees; it comprises of 13 protein-coding, 24 putative tRNA, and two rRNA genes, along with an A + T-rich control region. However, the number of tRNA-Met (three) in A. koschevnikovi was two more than the number present in other cavity-nesting honeybees. Similar to other honeybee mitochondrial genomes, the heavy strand (H-strand) was predicted to contain nine protein-coding genes and 16 tRNA genes. The light strand (L-strand) was predicted to contain four protein-coding, eight tRNA, and two rRNA genes. All the tRNA genes possessed cloverleaf secondary structures. The ATP6 and ATP8 genes shared 19 nucleotides. Eight protein-coding genes of the A. koschevnikovi mitochondrial genome started with ATT, and ATP6, COIII, ND4 and Cytb genes started with ATG. The stop codon of each of these protein-coding genes was either TAA or TAG, as observed in the case of other honeybees.
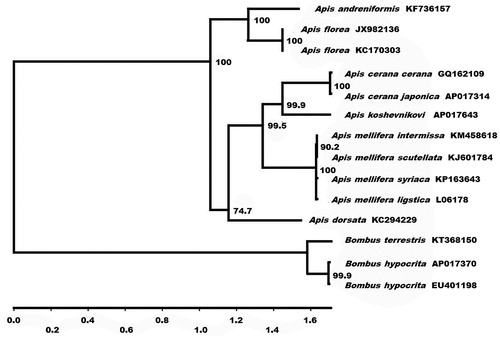
Phylogenetic analysis was performed using the sequences of 13 mitochondrial protein-coding genes and those of 14 closely related taxa (). The phylogenetic analyses of the complete mitochondrial DNA genes strongly supported the result obtained from the phylogenetic analysis of partial DNA sequences (Arias & Sheppard Citation2005), grouping the monophyletic species within the cavity-nesting honeybees: A. mellifera (Crozier & Crozier Citation1993; Haddad Citation2015; Gibson & Hunt Citation2016; Hu et al. Citation2016), A. koschevnikovi, and A. cerana (Tan et al. Citation2011; Takahashi et al. Citation2016). The complete sequence of the A. koschevnikovi mitochondrial genome provides additional genetic tools for studying conservational genetics and biogeography of this species.
Figure 1. Phylogenetic relationships (maximum likelihood) of the genus Apis and Bombus of among the members of Apinae based on the nucleotide sequence of 13 protein-coding genes regions in the mitochondrial genome. The numbers beside the nodes are percentages of 1000 bootstrap values. The Bombus terrestris and B. hypocrita were used as an outgroup. Alphanumeric terms indicate the GenBank accession numbers.
Acknowledgements
We are grateful to Prof. Tadaharu Yoshida and Prof. Masami Sasaki (Tamagawa University) for kindly providing honeybee sample.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Arias MC, Tingek S, Kelitu A, Sheppard WS. 1996. Apis nuluensis Tingek, Koeniger and Koeniger, 1996. and its genetic relationship with sympatric species inferred from DNA sequences. Apidologie. 27:415–422.
- Arias MC, Sheppard WS. 2005. Phylogenetic relationships of honey bees (Hymenoptera:Apinae:Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol Phylogenet Evol. 37:25–35.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol Phylogenet Evol. 69:313–319.
- Crozier RH, Crozier YC. 1993. The mitochondrial genome of the honeybee Apis mellifera: Complete sequence and genome organization. Genetics. 133:97–117.
- Gibson JD, Hunt GJ. 2016. The complete mitochondrial genome of the invasive Africanized Honey Bee, Apis mellifera scutellata (Insecta: Hymenoptera: Apidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:561–562.
- Haddad NJ. 2015. Mitochondrial genome of the Levant Region honeybee, Apis mellifera syriaca (Hymenoptera: Apidae). Mitochondrial DNA. 30:1–2.
- Hadisoesilo S, Raffiudin R, Susanti W, Atmowidi T, Hepburn C, Radloff SE, Fuchs S, Hepburn HR. 2008. Morphometric analysis and biogeography of Apis koschevnikovi Enderlein (1906). Apidologie. 39:495–503.
- Hu P, Lu ZX, Haddad N, Noureddine A, Loucif-Ayad Q, Qang YZ, Zhao RB, Zhang AL, Guan X, Zhang HX, Niu H. 2016. Complete mitochondrial genome of the Algerian honeybee, Apis mellifera intermissa (Hymenoptera: Apidae). Mitochondrial DNA. 27:1791–1792.
- Jobb G, von Haeseler A, Strimmer K. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 4:18.
- Takahashi J, Wakamiya T, Kiyoshi T, Uchiyama H, Yajima S, Kimura K, Nomura T. 2016. The complete mitochondrial genome of the Japanese honeybee, Apis cerana japonica (Insecta: Hymenoptera: Apidae). Mitochondrial DNA Part B. 1:156–157.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tan HW, Liu GH, Dong X, Lin RQ, Song HQ, Huang SY, Yuan ZG, Zhao GH, Zhu XQ. 2011. The complete mitochondrial genome of the Asiatic cavity-nesting honeybee Apis cerana (Hymenoptera: Apidae). PLoS One. 6:e23008.
- Tanaka H, Roubik DW, Kato M, Liew F, Gunsalam G. 2001. Phylogenetic position of Apis nuluensis of northern Borneo and phylogeography of A. cerana as inferred from mitochondrial DNA sequences. Insectes Soc. 48:44–51.
- Tanaka H, Suka T, Kahono S, Samejima H, Mohamed M, Roubik DW. 2003. Mitochondrial variation and genetic differentiation in honey bees (Apis cerana, A. koschevnikovi and A. dorsata) of Borneo. Tropics. 13:107–117.
