Abstract
Here, we present the first complete mitochondrial genome of the Red Tailed Loach Yasuhikotakia modesta (Teleostei: Botiidae) from Thailand, assembled from next-generation transcriptome sequencing data. The assembled transcript corresponds to the full length mitochondrial genome of Y. modesta, which measured 16,865 bp in length, and contained 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. A slight A + T bias was observed in the mitogenome of Y. modesta with an overall base composition of 32.2% A, 25.8% T, 26.4% C, and 15.4% G, and a GC content of 41.8%. The gene arrangement was identical to that of previously described loach mitogenomes.
Freshwater fishes of the family Botiidae contain eight genera and 80 species which are widespread across South-, Southeast-, and East Asia. They are high-priced for their taste and a number of species, especially from the genera Botia, Chromobotia, and Yasuhikotakia are famous in ornamental fish keeping. One of the two subfamilies of Botiidae, the Botiinae, contains only tetraploid species, but details concerning this polyploidization remain unknown (Šlechtová et al. Citation2006). The reconstruction of a reliable phylogeny of Botiidae will be best achieved with the use of genetic data, including mitochondrial genes. Here, we present the full mitogenome of Y. modesta, a tetraploid species that occurs in the Mekong River and Chao Phraya River (Nalbant Citation2002).
The specimen was collected near the Mekong mainstream at the locality of Ubon Ratchathani (15 20′16″N 105 28′00″E) and deposited in the collection of the Institute of Animal Physiology and Genetics in Liběchov, Czech Republic, under the accession number A9732.
A full-length transcript of the mitogenome was obtained from the next-generation transcriptome sequencing. Paired end Illumina sequencing libraries were generated from 600ng total RNA input with fragment sizes of 260 bp as described by Gao et al. (Citation2014). The libraries were sequenced on an Illumina HiSeq2000 platform using Illumina RNA TruSeq protocol and kit v2. Sequencing yielded 117,040,239 100 bp paired end reads, which were used for a de novo transcriptome assembly of Y. modesta with SOAPtransdenovo2 (v240) (Xie et al. Citation2014) and parameters -K 31 -M 3 -F. The assembled mitogenome transcript was manually inspected for repeats at the transcript ends to confirm circularity. Annotations were carried out with MITOchondrial genome annotation Server (MITOS) (Bernt et al. Citation2012), and manual validation of the coding regions using the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
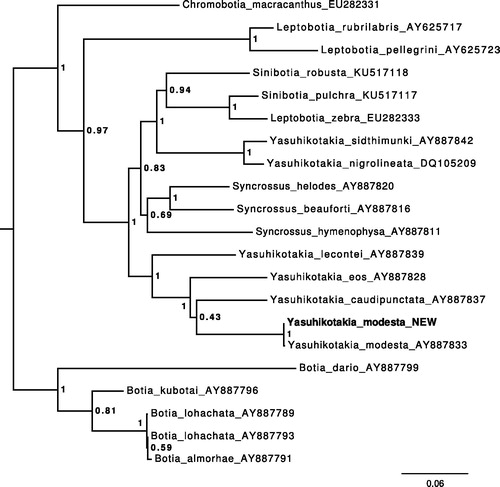
The annotated sequence file was submitted to NCBI (accession no. KY131962). The phylogenetic position of the new sequence of Y. modesta according to the gene Cytochrome B is shown in .
Figure 1. Maximum-likelihood tree illustrating the phylogenetic position of the newly sequenced Y. modesta gene sequence among a subset of Botiidae species. Cytochrome B sequences were aligned using MAFFT 7.271 and highly divergent or poorly aligned regions were removed with Gblocks 0.91b (Castresana Citation2000) allowing for gap positions and smaller blocks. Trees were calculated using PhyML 3.1 (Guindon et al. Citation2010) with 12 rate categories, optimized equilibrium frequencies, GTR model of sequence evolution and combined heuristics (nearest neighbour interchange and subtree pruning and rerafting).
The complete mitochondrial transcript of Y. modesta was 16,865 bp in length and contained 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, and 22 transfer RNA genes. As described for other fish mitogenomes (Yu et al. Citation2016), the mitochondrial genome of Y. modesta contained a slight A + T bias with an overall base composition of 32.2% A, 25.8% T, 26.4% C, and 15.4% G. The gene arrangement of the present mitogenome is similar to those of other loaches (Wang et al. Citation2012; Tang et al. Citation2013). Most of the genes were encoded on the L-strand with the exception of ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer2, tRNAGlu, and tRNAPro) which are encoded in the H-strand. All PCGs had ATG as initiation codon with the exception of CYTB, which used GTG as initiation codon. TAA was the most used termination codon with the exception of COX3, CYTB, ND2, ATP8, and ND3, which used a TAG termination codon. The 12S and 16S genes had a length of 947 and 1673 bp, respectively.
Disclosure statement
This study was funded by the Grant Agency of the Czech Republic, grant number 13 - 37277 S. The authors declare no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2012. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet E. 7:332–336.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Gao J, Schatton D, Martinelli P, Hansen H, Pla-Martin D, Barth E, Becker C, Altmueller J, Frommolt P, Sardiello M, Rugarli EI. 2014. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J Cell Biol. 207:213–223.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Nalbant TT. 2002. Sixty million years of evolution. Part one: family Botiidae (Pisces: Ostariophysi: Cobitoidea). Trav Mus Natl Hist Nat Grigore Antipa. 44:309–333.
- Šlechtová V, Bohlen J, Freyhof J, Ráb P. 2006. Molecular phylogeny of the Southeast Asian freshwater fish family Botiidae (Teleostei: Cobitoidea) and the origin of polyploidy in their evolution. Mol Phylogenet Evol. 39:529–541.
- Tang Q, Huang Y, Wang J, Huang J, Wang Z, Peng Z. 2013. The complete mitochondrial genome sequence of Triplophysa bleekeri (Teleostei, Balitoridae, Nemacheilinae). Mitochondrial DNA. 24:25–27.
- Wang J, Tang Q, Wang Z, Zhang Y, Wu Q, Peng Z. 2012. The complete mitogenome sequence of a cave loach Triplophysa rosa (Teleostei, Balitoridae, Nemacheilinae). Mitochondrial DNA. 23:366–368.
- Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, Zhou X. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics. 30:1660–1666.
- Yu P, Wei M, Yang Q, Yang Y, Wan Q. 2016. The Complete Mitochondrial Genome of Ambastaia sidthimunki (Cypriniformes: Cobitidae). Mitochondrial DNA. Part A. 27:3228–3229.
