Abstract
In this study, we report the complete mitochondrial genome of Sinocyclocheilus ronganensis. The whole mitochondrial genome is 16,587 bp with an accession number KX778473, and consists of 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs genes, and a 936 bp control region. Phylogenetic analysis shows that S. ronganensis is close to cave-restricted S. anophthalmus and surface-dwelling S. grahami. The complete mitogenome of S. ronganensis may provide useful information for studying the genetic mechanism of cavefish, and enrich the fish mitochondrial genome resource for further research.
Sinocyclocheilus ronganensis, a cavefish and is newly found in the karst river of Rong’an county, Southwestern China (Luo et al. Citation2016). Like other surface-dwelling species (Zhao & Zhang Citation2009; Meng et al. Citation2013), S. ronganensis has normal eyes, full-cover scales but larger than most of them, and it is a good model for studying the genetic status and evolutionary by comparing the mitochondrial genome.
The S. ronganensis sample was collected from subterranean river of Rong’an county (24°59′59″N, 109°28′02″E), Southwestern China in March, 2016, and the specimen was stored in Liuzhou Aquaculture Technology Extending Station, Liuzhou, China. In this study, the complete mtDNA was sequenced using the Illumina Hiseq4000 platform with de novo strategy, and then the phylogenetic tree was established using the neighbour-joining (NJ) method.
The complete mitochondrial genome of S. ronganensis had been deposited in the GenBank with an accession number KX778473. The mitochondrial genome was 16,587 bp in length, with the base composition of 31.57% A, 25.41% T, 26.82% C, 16.21% G, and an AT bias of 56.97%. Besides, it consisted of 13 protein-coding, 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and 1 control region (D-loop). So far, the arrangement of these genes was identical to that found in the species of Sinocyclocheilus (Wu et al. Citation2010; He et al. Citation2016; Peng et al. Citation2017). Except for ND6 gene and eight tRNA genes, all other genes were located on the heavy strand (H-strand). All the 13 protein-coding genes contained the same start codon ATG except the gene COXI, which contained GTG instead, however, the termination codons of the 13 protein-coding genes were different, with either TAA, T––, TA–, or TAG. 22 tRNA genes were ranged from 67 to 76 bp in size, and one 12S rRNA and one 16S rRNA with corresponding sizes of 954 and 1680 bp, respectively. A total of 936 bp control region (D-loop) was located between tRNA-Phe and tRNA-Pro. In all genes, the overlaps and spaces might be existed in adjacent gene, for example, tRNA-Asn (32 bp spaces), tRNA-Asp (13 bp spaces), ATP8 (7 bp overlaps), ND4L (7 bp overlaps) were found obviously.
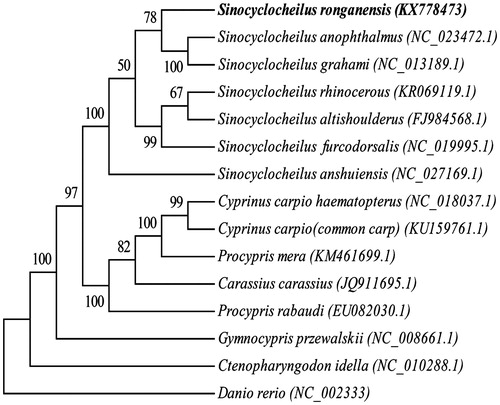
Sinocyclocheilus ronganensis and S. grahami had the same lift-style, and also had the same normal eyes and scales, but interesting, in our phylogenetic tree, the surface-dwelling S. grahami and the cave-restricted S. anophthalmus first clustered together and later this group clustered with S.ronganensis (), it suggested that phenotypic convergence might be caused by similar environmental conditions despite the difference in mitochondrial evolution of Sinocyclocheilus, and this results of this study was also in agreement with previous reports (Xiao et al. Citation2005; He et al. Citation2016; Peng et al. Citation2017).
Disclosure statement
The authors report no conflicts of interest and are responsible for the content and writing of the paper. This work was supported by Liuzhou Fiscal Fund 2015(No. 54), and Special Fund for Agro-scientific Research in the Public Interest of the People’s Republic of China (No. 201303048).
Additional information
Funding
References
- He SY, Lu J, Jiang WS, Yang S, Yang JX, Shi Q. 2016. The complete mitochondrial genome sequence of a cavefish Sinocyclocheilus anshuiensis (Cypriniformes: Cyprinidae). Mitochondrial DNA. 27:4256--4258.
- Luo FG, Huang J, Liu X, Luo T, Wen YH. 2016. Sinocyclocheilus ronganensis Luo, Huang et Wen sp. nov., a new species belonging to Sinocyclocheilus Fang from Guangxi (Cypriniformes: Cyprinidae). J South Agric. 47:650–655. in Chinese with English abstracts.
- Meng F, Braasch I, Phillips JB, Lin X, Titus T, Zhang C, Postlethwait JH. 2013. Evolution of the eye transcriptome under constant darkness in Sinocyclocheilus cavefish. Mol Biol Evol. 30:1527–1543.
- Peng S, Lu J, Yang JX, Chen XL, Shi Q. 2017. The complete mitochondrial genome of horned Golden-line barbell, Sinocyclocheilus rhinocerous (Cypriniformes, Cyprinidae). Mitochondrial DNA. 28: 269--270.
- Wu XJ, Wang L, Chen SY, Zan RG, Xiao H, Zhang YP. 2010. The complete mitochondrial genomes of two species from Sinocyclocheilus (Cypriniformes: Cyprinidae) and a phylogenetic analysis within Cyprininae. Mol Biol Rep. 37:2163–2171.
- Xiao H, Chen SY, Liu ZM, Zhang RD, Li WX, Zan RG, Zhang YP. 2005. Molecular phylogeny of Sinocyclocheilus (Cypriniformes: Cyprinidae) inferred from mitochondrial DNA sequences. Mol Phylogenet Evol. 36:67–77.
- Zhao YH, Zhang CG. 2009. Endemic fishes of Sinocyclocheilus (Cypriniformes: Cyprinidae) in China–species diversity, cave adaptation, systematics and zoogeography. Beijing: Science Press.