Abstract
The complete mitochondrial genome of Dabry’s sturgeon (Acipenser dabryanus) was first determined in this study. The mitochondrial DNA is circular molecule (16,437 bp in length) with a typical vertebrate mitochondrial gene arrangement, including 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 1 non-coding control region (D-loop). All genes were encoded on the heavy strain except for ND6 and eight tRNA genes. The overall base composition of the heavy strain was A (30.2%), T (23.7%), C (29.7%), G (16.4%), and with an AT bias of 53.9%. Phylogenetic analysis based on the mitochondrial genomes of 10 related species using MEGA 5.1 indicated that A. dabryanus and A. sinensis are the most closely related species.
Dabry’s sturgeon (Acipenser dabryanus), belonging to the family Acipenseriforms, Acipenseridae, is a Critically Endangered and endemic species which has been distributed in the upper and middle sections of the Yangtze River and its large tributaries (Que et al. Citation2014). However, the natural populations have declined sharply in the past decades due to dam construction, overfishing, pollution, etc., in the Yangtze River. Thus, A. dabryanus has been listed as Class I-Protected species in China (Zhang et al. Citation2011).
The sampled specimen was collected from Xiangjiaba breeding and releasing station (N 28°37′45.70″, E 104°25′41.70″) in January 2016, China. This specimen was stored in Institute of Chinese Sturgeon with the accession number: YC20160118F02. Total genomic DNA was extracted from caudal fins by a traditional phenol-chloroform method (Sambrook & Russell Citation2002). Eight pairs of PCR primers were designed for amplification of the mitogenome sequence. The complete mitogenome of A. dabryanus (GenBank accession number NC005451) is 16,437 bp in length, including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 1 non-coding control region. This gene arrangement of Dabry’s sturgeon is similar to the typical vertebrate mitogenomes (Broughton & Reneau Citation2006). All genes were encoded on the heavy strain except for ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro). The overall base composition of the heavy strain was A (30.2%), T (23.7%), C (29.7%), G (16.4%), with a slight bias to AT (53.9%). Twelve of the protein-coding genes share the start codon ATG, except for COI, which begins with GTG. Ten protein-coding genes use TAA or TAG as stop codon, although three of the genes (COII, ND4, Cytb) require T–– or TA– as incomplete stop codon.
Figure 1. Neighbor-joining molecular phylogenetic tree of 10 species of Acipenseriforms based on 13 protein-coding gene sequences. Numbers at each branch is 1000 bootstrap value.
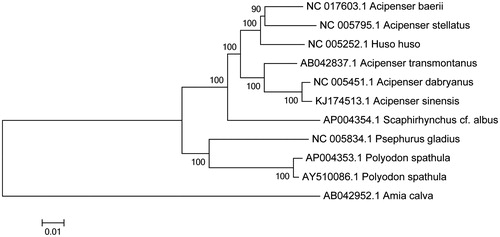
The mtDNA sequences of 10 species of fishes were downloaded from GenBank, Amia calva as an out-group. Phylogenetic analysis based on the mitochondrial genomes of 10 related species by using MEGA 5.1 (Kumar et al. Citation2008) indicated that A. dabryanus and A. sinensis are the most closely related species ().
Disclosure statement
The authors report no conflicts of interest. This study was supported by the China Three Gorges Corporation program (No. 2013-Z11025).
Additional information
Funding
References
- Broughton RE, Reneau PC. 2006. Spatial covariation of mutation and nonsynonymous substitution rates in vertebrate mitochondrial genomes. Mol Biol Evol J. 23:1516–1524.
- Kumar S, Nile M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformatics. 9:299–306.
- Que Y, Xu D, Shao K, Xu N, Li W, Zhu B. 2014. Identification and characterization of seventeen novel microsatellite markers for Dabry’s sturgeon (Acipenser dabryanus). J. Genet. 93:62–65.
- Sambrook J, Russell DW. 2002. Molecular cloning: a laboratory manunal. 3rd ed. Beijing: Science Press.
- Zhang H, Wei Q, Du H, Li LX. 2011. Present status and risk for extinction of the Dabry’s sturgeon (Acipenser dabryanus) in the Yangtze River watershed: a concern for intensified rehabilitation needs. J Appl Ichthyol. 27:181–185.
