Abstract
Complete mitochondrial genome of Phytophthora infestans, A2 mating type (MT) with a size of ≅37,767 bp was sequenced. A total of 53 protein-coding genes are predicted on both strands, including 25 tRNA, 2 rRNA, and 18 respiratory proteins. Gene order of A2MT was consistent with that established in A1, despite high level of polymorphism in both coding and non-coding regions. The mtDNA of A2MT was found to have 99.5% and 99.4% homology with Ia and Ib, whereas 94.7% and 94.3% with IIa and IIb, respectively. Study of repeats revealed a dinucleotide (AT)9 specific to A1 and homology of cox1 gene sequence revealed the relationship among 50 Phytophthora species.
Ever since triggering the Irish famine, Phytophthora infestans (Mont.) has continued to wreak havoc on potato fields throughout world and its population has undergone drastic changes with new population detected, having more pathotypes, carrying new MT and being resistant to metalaxyl (Chimote et al. Citation2010; Arora et al. Citation2014). Since past two decades, it is reported to be heterothallic, resulting in high level of genetic variation and rapid evolution (Singh et al. Citation1994). Population displacement by genotypes with increased fitness is a recurrent event, for instance in India both MTs exists, with A2MT completely replacing A1MT in the hills and almost stabilizing, while in the plains A1MT has established itself (Chimote et al. Citation2010; Arora et al. Citation2014). Polymorphism at various regions of mitochondrial-genome and even complete mitochondrial-genome of P. infestans has been successfully employed to study origin, migration, and diversity (Hwang et al. Citation2014). To date mtDNA of A1MT have been sequenced, revealing the phylogenetic relationship among haplotype I(a&b) and II(a&b), but no efforts have been made to excavate relationship existing between the MTs(A1/A2). In the present study, complete mitochondrial-genome sequence of A2MT is reported and compared with A1MTs along with three related Phytophthora species.
P. infestans (HP10-31),1 belonging to haplotype-Ia (Carter et al. Citation1990) and A2MT, were isolated from late blight-infected potato fields in Shimla, placed in mid-Himalayas with wet temperate climate (31.61°N,77.10°E). The culture was grown and maintained (Caten & Jinks Citation1968), harvested mycelium was crushed and dispersed in pre-cooled sucrose buffer. The nuclei and mitochondria were separated by differential centrifugation (Klimczak & Prell Citation1984) and DNA was isolated by CTAB (Murray & Thompson Citation1980). The mtDNA was sheared; shotgun library was prepared (Roche-Diagnostics) and sequenced using gsFLX_Titanium (Roche-454), and 40Mb data was obtained with coverage of ≅100×. The sequence data was assembled using GS_DeNova_Assember and GS_Ref_Mapper (Roche-Diagnostics) with mitochondrial-genome of Ia as reference, yielding single mega scaffold of 37,767bp covering entire genome with 22.38% GC content. A total of 55 protein-coding genes were predicted using mVISTA, including 26 tRNA (tRNAScan-SE), 16 ribosomal proteins (RNAmmer_v1.2), 18 respiratory proteins, and an import protein Sec-Y (independent transport protein). None of the predicted genes possessed introns, ATG is the start codon for all the genes whereas TAA is the stop codon for all the genes except nadh11 which has TGA (Korkmaz et al. Citation2014). MISA studies revealed the presence of only Di-and Tri-nucleotide repeats among all Phytophthora species. Two Di-nucleotide motifs (AT)7 and (AT)6 were found to be common in both MTs, whereas motif (AT)15 was found specific to A2MT, further exploited for differentiating MTs. The A2 mating type formed close cluster with haplotype Ia (0.995) and Ib (0.994) of A1 mating type in the phylogenic relationship using average nucleotide identity (ANI) (). Comparing complete sequence of cox1(1479 bp) gene for 50 Phytophthora species using CLUSTALW revealed that P. infestans, P. iranica, P. mirabilis, P. clandestina, P. andina, P. ipomeae and P. phaseoli formed a single cluster indicating the close relations between species (Martin & Tooley Citation2003; Kroon et al. Citation2004). The remaining Phytophthora species formed a bigger cluster, whereas P. aphanidermatum and P. brassicae formed unexpectedly separate branches (Kroon et al. Citation2004).
Figure 1. The phylogenetic tree depicting Genome-wide comparative studies of A2 MT with A1 MTs, P. ramorum, P. sojae and P. andina using Average Nucleotide Identity http://enve-omics.ce.gatech.edu/ani/. P. infestans A1 MT has 99.5%, 94.2%, 99.4%, and 94.3% sequence similarity with Ia (Acc. No. AY894835), IIa (Acc. No. AY898627), Ib (Acc. No. NC002387), and IIb (Acc. No. AY898628), respectively, and 98.3%, 71.5%, and 67.5% sequence similarity with P. andina (Acc. No. NC015619), P. ramorum (Acc. No. NC009384), and P. sojae (Acc. No. NC009385), respectively. Several nuclear and mitochondrial gene studies have shown that P. andina is an hybrid, with P. infestans as one of the parents (Kroon et al. Citation2004; Gómez-Alpizar et al. Citation2008; Haas et al. Citation2009; Goss et al. Citation2011; Blair et al. Citation2012; Lassiter et al. Citation2015) and our results strongly support the same at whole-genome level.
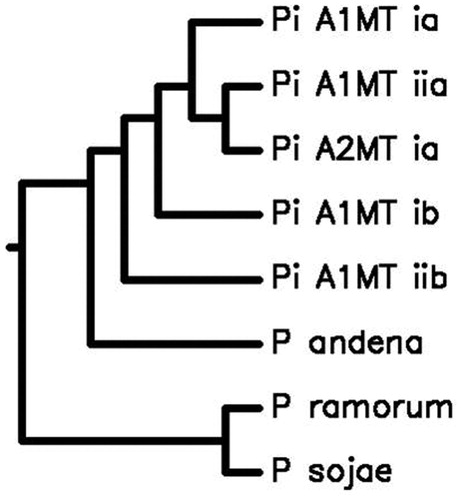
The high diversity found between mitochondrial-genomes of two MTs may not be only due to host specialization among the pathogens leading to evolution of novel mitochondrial lineages, but may be due to the migration dynamics and the climatic variations. The hypothesis is further supported by the increase in the GC content of the mt genome of A2MT. However, before firm conclusions about the genome variability between the MTs can be drawn, additional comparisons among more genotypes are needed to clarify this.
Nucleotide sequence accession no.
Complete mitochondrial genome sequence of P. infestans A2 mating type (Haplotype Ia) is submitted to NCBI/GeneBank under the accession no. KU837230, and first draft sequence of whole genome has been submitted under Acc. No. LYVM00000000, Version: LYVM01000000.
| Abbreviations | ||
| MT | = | Mating type |
| mt | = | Mitochondria |
| CTAB | = | Cetyltrimethylammonium bromide |
| ANI | = | Average nucleotide identity |
| MISA | = | MIcroSAtellite identification tool |
Acknowledgements
The authors highly knowledge the contribution of Youvika Singh and Shashi Rawat for assisting the whole genome assembly and data submission through CABin project at Central Potato Research Institute, Shimla.
Disclosure statement
None of the authors report any conflict of interest. The authors alone are responsible for the content and writing of the paper.
Note
Additional information
Funding
Notes
1 The specimen P. infestans Phylotype Ia and A2 Mating Type has been submitted to gene bank for agriculturally important microbes at ICAR-Central Potato Research Institute, Shimla with No. HP10-31.
References
- Arora RK, Sharma S, Singh BP. 2014. Late blight disease of potato and its management. Potato J. 41:16–40.
- Blair JE, Coffey MD, Martin FN. 2012. Species tree estimation for the late blight pathogen, Phytophthora infestans, and close relatives. PLoS One. 7:e37003.
- Carter DA, Archer SA, Buck KW. 1990. Restriction fragment length polymorphism of mitochondrial DNA of Phytophthora infestans. Mycol Res. 8:1123–1128.
- Caten CE, Jinks JL. 1968. Spontaneous variability of single isolates of Phytophthora infestans cultural variation. Can J Bot. 46:329–347.
- Chimote VP, Kumar M, Sharma PK, Singh PH, Singh BP. 2010. Characterization of changes in phenotype and genotype of Phytophthora infestans isolates from India. J Plant Pathol. 92:669–677.
- Gómez-Alpizar L, Hu CH, Oliva RF, Forbes GA, Ristaino JB. 2008. Phylogenetic relationships of Phytophthora andina, a new species from the highlands of Ecuador that is closely related to the Irish potato famine pathogen Phytophthora infestans. Mycologia. 100:590–602.
- Goss EM, Cardenas ME, Myers K, Forbes G, Fry WE, Restrepo S, Grünwald N. 2011. The plant pathogen Phytophthora andina emerged via hybridization of an unknown Phytophthora species and the Irish potato famine pathogen, P. infestans. PLoS One. 6:e24543.
- Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Alalibo TT, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 461:393–398.
- Hwang YT, Wijekoon C, Kalischuk M, Johnson D, Howard R, Prufer D, Kawchuk L. 2014. Evolution and management of the Irish Potato famine pathogen Phytophthora infestans in Canada and the United States. Am J Potato Res. 91:579.
- Klimczak LJ, Prell HH. 1984. Isolation and characterization of mitochondrial DNA of the oomycetous fungus Phytophthora infestans. Curr Genet. 8:323–326.
- Korkmaz G, Holm M, Wiens T, Sanyal S. 2014. Comprehensive analysis of stop codon usage in bacteria and its correlation with release factor abundance. J Biol Chem. 289:30334–30342.
- Kroon LPNM, Bakker FT, van der Bosch GBM, Bonants PJM, Flier WG. 2004. Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet Biol. 41:766–782.
- Lassiter ES, Russ C, Nusbaum C, Zeng Q, Saville A, Olarte R, Carbone I, Hu C-H, Seguin-Orlando A, Samaniego JA, et al. 2015. Mitochondrial genome sequences reveal evolutionary relationships of the Phytophthora 1c clade species. Curr Genet. 61:567–577.
- Martin FN, Tooley PW. 2003. Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia. 95:269–284.
- Murray HG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321–4325.
- Singh BP, Roy S, Bhattacharyya SK. 1994. Occurrence of the A2 mating type of Phytophthora infestans in India. Potato Res. 37:227–231.
