Abstract
The yellow-legged Asian hornet, Vespa velutina nigrithorax, which originated from Asia, has invaded several countries, including South Korea. In Korea, V. velutina nigrithorax predation on honeybees is one of the most serious factors threatening apiculture. We sequenced the complete mitochondrial genome (mitogenome) of V. velutina to better understand the mitogenomic characteristics of this species. The 16,475 bp mitogenome of V. velutina consists of a typical set of genes, with an arrangement identical to that of congeneric species. Vespa velutina possesses the shortest A + T-rich region (132 bp) among congeneric Vespa, and this is also the shortest in the superfamily Vespoidea. Phylogenetic analysis using the 13 protein-coding genes of Vespoidea species indicated that each family forms strongly supported monophyletic groups (Bayesian posterior probability =1; ML, 100%). Moreover, V. velutina and V. bicolor form strongly supported sister groups (Bayesian posterior probability =1; ML, 100%).
Originally distributed from northern India to the Indochinese Peninsula, Taiwan, and Indonesia (Rortais et al. Citation2010) the yellow-legged Asian hornet, Vespa velutina nigrithorax, has invaded Europe and also South Korea (Haxaire et al. Citation2006; Kim et al. Citation2006; Castro & Pagola-Carte Citation2010; Bruneau Citation2011; Villemant et al. Citation2011). First observed in Korea in 2003 (Kim et al. Citation2006), the species has now spread to nearly all regions of South Korea, the sole exception being the remote island of Jeju. This species is one of the major apicultural pests, and is also a pest in urban areas. During summer, the economic losses caused by this devastating bee-hawking hornet are so severe that beekeeping becomes almost unviable.
In this study, we sequenced the complete mitochondrial genome (mitogenome) of V. velutina to better understand the mitogenomic characteristics of this species and its phylogenetic relationships within Vespoidea. In 2016, one adult V. velutina was captured at Juam-myeon, in Jeollanamdo Province in Korea (35°04′16.1″N, 127°13′38.8′′E) using a commercial hornet trap. This voucher specimen was deposited in Chonnam National University, Gwangju, Korea, under the accession no. CNU6596.
Using total DNA as a template, three long overlapping fragments (COI–ND4, ND4-lrRNA, and lrRNA–COI) were amplified, and subsequently, 32 short overlapping fragments were amplified using the three long fragments as templates. The primers used in the present study for amplifying the long- and short-fragments were designed from available Vespa mitogenomes (Chen et al. Citation2016; Wei et al. Citation2016).
The V. velutina mitogenome is 16,475 bp in size and includes the typical sets of genes (2 rRNAs, 22 tRNAs, and 13 protein-coding genes [PCGs]) and a major non-coding A + T-rich region (GenBank accession number KY091645). At 16,476 bp, the mitogenome of V. velutina is larger than other Vespa mitogenomes that have been completely sequenced to date (15,779 bp in V. ducalis, Kim et al., In Press; 15,902 bp in V. mandarinia, Chen et al. Citation2016). The gene arrangement in the V. velutina mitogenome is identical to that of other Vespa species, but it differs substantially from the ancestral insect order found in the majority of insects (Boore Citation1999).
The A/T content among genes and regions varies markedly in the V. velutina mitogenome: 92.4% in the A + T-rich region, 86.0% in tRNAs, 85.6% in lrRNA, 84.4% in srRNA, and 79.7% in PCGs. All V. velutina PCGs begin with the typical ATN codon (six with ATG, five with ATT, one with ATC, and one with ATA). Twelve of the 13 PCGs end with TAA, whereas COIII ends with a single T. The incomplete termination codon results in a complete TAA stop codon via post-translational modifications occurring during the mRNA maturation process (Ojala et al. Citation1981).
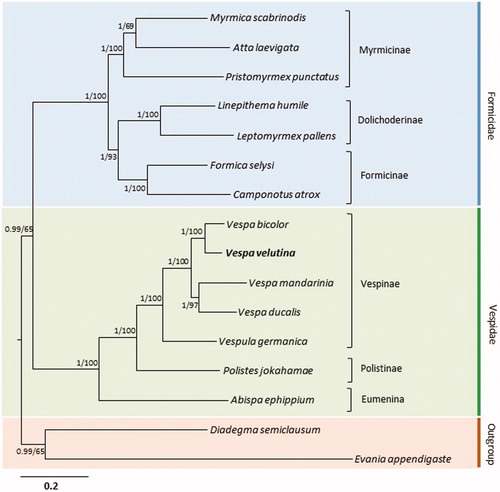
We performed phylogenetic analysis using the 13 PCGs of 14 mitogenome sequences from Vespoidea, including that of V. velutina, with the inclusion of one species each from Ichneumonidae and Evaniidae belonging to Vespoidea as outgroups (Wei et al. Citation2009, Citation2010). Both Bayesian inference (BI) and maximum-likelihood (ML) methods (Stamatakis, Citation2006; Ronquist et al. Citation2012) were performed using the GTR + GAMMA + I model in CIPRES Portal v. 3.1 (Miller et al. Citation2010). Using these two analyses, identical topologies were obtained, with both Formicidae and Vespidae forming strongly supported monophyletic groups (; Bayesian posterior probability = 1; ML, 100%). Within Vespa, the sister relationships between V. ducalis and V. bicolor and between V. mandarinia and V. ducalis were strongly supported (BPP = 1; ML, 97–100%). Previous phylogenetic analysis of Vespa based on combined analyses of morphological (45 characters) and molecular (four mt genes and two nuclear genes) data also support the close relationships between V. ducalis and V. bicolor and between V. mandarinia and V. ducalis (Perrard et al. Citation2013).
Figure 1. Phylogeny of Vespoidea. Bayesian Inference (BI) and maximum-likelihood (ML) methods produced the same topology based on 13 concatenated protein-coding genes. The numbers at each node specify percentage Bayesian posterior probabilities generated by BI analysis (first value) and bootstrap percentages of 1000 pseudo-replicates generated by ML analysis (second value). The scale bar indicates the number of substitutions per site. One species from the Ichneumonidae (Diadegma semiclausum) and one species from the Evaniidae (Evania appendigaste) were utilized as outgroups. GenBank accession numbers are as follows: V. ducalis, KX950825; V. mandarinia, KR059904; V. bicolor, KJ735511; Vespula germanica, KR703583; Polistes jokahamae, KR052468; Abispa ephippium, EU302588; Atta laevigata, KC346251; Myrmica scabrinodis, LN607806; Pristomyrmex punctatus, AB556946; Camponotus atrox, KT159775; Formica selysi, KP670862; Linepithema humile, KX146468; Leptomyrmex pallens, KC160533; D. semiclausum, EU871947; and E. appendigaste, FJ593187.
Disclosure statement
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Agri-Bio Industry Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (316038-3).
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Bruneau E. 2011. Le frelon asiatique, déjà là. ActuApi. 55:1–6.
- Castro L, Pagola-Carte S. 2010. Vespa velutina Lepeletier, 1836 (Hymenoptera: Vespidae) recolectada en la Península Ibérica. Heteropterus Revista De Entomología. 10:193–196.
- Chen PY, Wei SJ, Liu JX. 2016. The mitochondrial genome of the Vespa mandarinia Smith (Hymenoptera: Vespidae: Vespinae) and a phylogenetic analysis of the Vespoidea. Mitochondrial DNA Part A. 27:4414–4415.
- Haxaire J, Bouguet J-P, Tamisier J-P. 2006. Vespa velutina Lepeletier, 1836, une redoutable nouveauté pour la faune de France (Hymenoptera, Vespidae). Bulletin De La Société Entomologique De France. 111:194.
- Kim JK, Choi MB, Moon TY. 2006. Occurrence of Vespa velutina Lepeletier from Korea, and a revised key for Korean Vespa species (Hymenoptera: Vespidae). Entomol Res. 36:112–115.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proc Gateway Comput Environ Workshop (GCE), New Orleans, p. 1–8.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290:470–474.
- Perrard A, Pickett KM, Villemant C, Kojima J, Carpenter JM. 2013. Phylogeny of hornets: a total evidence approach (Hymenoptera, Vespidae, Vespinae, Vespa). J Hymenopt Res. 32:1–15.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Rortais A, Villemant C, Gargominy O, Rome Q, Haxaire J, Papachristoforou A, Arnold G. 2010. A new enemy of honeybees in Europe: the Asian hornet Vespa velutina. In: Settele J, ed. Atlas of biodiversity risks – from Europe to the globe, from stories to maps. Sofia & Moscow: Pensoft. p. 11.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Villemant C, Barbet-Massin M, Perrard A, Muller F, Gargominy O, Jiguet F, Rome Q. 2011. Predicting the invasion risk by the alien bee-hawking Yellow-legged hornet Vespa velutina nigrithorax across Europe and other continents with niche models. Biol Cons. 144:2142–2150.
- Wei SJ, Niu FF, Tan JL. 2016. The mitochondrial genome of the Vespa bicolor Fabricius (Hymenoptera: Vespidae: Vespinae). Mitochondrial DNA Part A. 27:875–876.
- Wei SJ, Shi M, He JH, Sharkey M, Chen XX. 2009. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome. 52:308–319.
- Wei SJ, Tang P, Zheng LH, Shi M, Chen XX. 2010. The complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A + T content and a long intergenic spacer between atp8 and atp6. Mol Biol Rep. 37:1931–1942.
