Abstract
The controversy surrounding the origin of antitropical distribution of Mytilus mussels and the taxonomic status of southern hemisphere populations remain unsolved, despite the efforts. One of the limiting factors remains the lack of the complete sequences of the representative mitochondrial genomes which would allow their proper comparison with the relatively well-represented northern hemisphere congeneric mussels. To fill this gap we sequenced the representative maternal (F) genome of a native Chilean mussel. The genome is 16,748bp long and structurally identical to the northern hemisphere M. edulis and M. galloprovincialis F genomes. However, the genetic distance from them (≈5%) is twice as high as the maximum distance between them (<2.5%). Thus, the notion that the name M. chilensis should be used for native Chilean Mytilus mussels, with the same rank as M. galloprovincialis and M. edulis is supported.
The Mytilus edulis species complex consists of three well-recognized marine mussels: M. edulis, M. trossulus, M. galloprovincialis and several populations found on the southern hemisphere (Hilbish et al. Citation2000). These southern populations have mixed ancestry, some are recently introduced by humans and some are native, with different distances from M. edulis and M. galloprovincialis of the northern hemisphere (Borsa et al. Citation2007). It has been shown that three separate mitochondrial lineages exist in southern hemisphere Mytilus spp. (Gérard et al. Citation2008). The status of these populations remains controversial. Some researchers call them either M. edulis or M. galloprovincialis of southern hemisphere (Westfall & Gardner Citation2010), while others use the term M. chilensis (Oyarzún et al. Citation2016). The officially accepted classification gives the native South American Mytilus mussels subspecies rank and the name M. edulis platensis d'Orbigny, 1842 and is supported by a critical review of literature (Borsa et al. Citation2012). It has been speculated that as many as four Mytilus species are present in the Magellane region of Chile (Oyarzún et al. Citation2016). Unfortunately, these are based on a limited number of nuclear and low resolution mitochondrial markers. Clearly, there is a need for better markers. Complete mitochondrial sequences contain enough information to precisely date the evolutionary events within the M. edulis species complex (Śmietanka et al. Citation2010), making them excellent marker candidates.
All Mytilus mussels exhibit an unusual system of mitochondrial DNA inheritance (Skibinski et al. Citation1994; Zouros et al. Citation1994). In this system two divergent mitochondrial genomes exist. However, the paternally transmitted M genome is present only in males whereas the maternally inherited F genome is more ubiquitous. Therefore, the F genome is more suitable for marker development.
Here we announce the complete sequence of the F mitochondrial genome from South American M. chilensis. It was isolated from a female specimen of Chilean Mytilus mussel sampled near Niebla, Valdivia (39° 51' 20” S, 73° 23' 35” W). It is representative for the local native population. The 16S sequence of this genome perfectly match native haplotypes (AM904582 and AM904585 from Gérard et al. Citation2008). The complete sequence was obtained in two steps: long range PCR, followed by re-amplifications and direct sequencing of shorter PCR products, as described previously (Śmietanka et al. Citation2010). The sequence has been deposited in GenBank under accession number KT966847 and the specimen is stored under CH1 accession number in our repository.
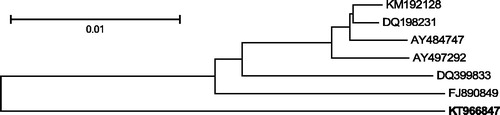
The number and order of genes in this genome is the same as in all other published mitochondrial F genomes from members of M. edulis species complex. There are also no major differences within the Control Region. The phylogenetic tree based on genetic distances between the newly sequenced genome and its closest published relatives was constructed (). The analysis confirms distinctiveness of southern hemisphere American Mytilus F genomes, supporting the view that M. chilensis should have the same taxonomic rank as M. galloprovincialis and M. edulis and should not be referred to as “southern hemisphere M. galloprovincialis” or “southern hemisphere M. edulis”.
Figure 1. Comparative analysis of the announced mitogenome. The following six complete F mitochondrial genomes, most similar (more than 94% sequence similarity) to the announced F genome of M. chilensis (KT966847, in bold) have been downloaded from GenBank: KM192128 and DQ198231 M. trossulus from the Baltic Sea (Zbawicka et al. Citation2007, Citation2014), AY484747 M. edulis from the Atlantic (Boore et al. Citation2004), AY497292 M. galloprovincialis from the Atlantic (Mizi et al. Citation2005), DQ399833 M. galloprovincialis from the Black Sea (Venetis et al. Citation2007), FJ890849 M. galloprovincialis from the Mediterranean Sea (Burzyński & Smietanka Citation2009). The sequences were aligned using MUSCLE (Edgar Citation2004). All alignment positions containing gaps and missing data were eliminated. There were a total of 16,247 positions in the final data set. The tree was inferred using the Neighbour-Joining method (Saitou & Nei Citation1987). The distances between sequences were computed as the number of base differences per site. The tree is drawn to scale, with branch lengths in the same units. The optimal tree, with the sum of branch lengths = 0.08 is shown. Bootstrap support (1000 replicates, Felsenstein Citation1985) was greater than 95% for all bipartitions. All analyses were conducted in MEGA6 (Tamura et al. Citation2013).
Acknowledgments
We thank Dr Karin Gérard for providing the samples and Ms Paulina Ulewska for help with laboratory work.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Boore JL, Medina M, Rosenberg LA. 2004. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the Scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol Biol Evol. 21:1492–1503.
- Borsa P, Daguin C, Bierne N. 2007. Genomic reticulation indicates mixed ancestry in southern-hemisphere Mytilus spp. mussels. Biol J Linn Soc. 92:747–754.
- Borsa P, Rolland V, Daguin-Thiébaut C. 2012. Genetics and taxonomy of Chilean smooth-shelled mussels, Mytilus spp. (Bivalvia: Mytilidae). C R Biol. 335:51–61.
- Burzyński A, Smietanka B. 2009. Is interlineage recombination responsible for low divergence of mitochondrial nad3 genes in Mytilus galloprovincialis? Mol Biol Evol. 26:1441–1445.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acid Res. 32:1792–1797.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791.
- Gérard K, Bierne N, Borsa P, Chenuil A, Féral J. 2008. Pleistocene separation of mitochondrial lineages of Mytilus spp. mussels from Northern and Southern Hemispheres and strong genetic differentiation among southern populations. Mol Phylogenet Evol. 49:84–91.
- Hilbish TJ, Mullinax A, Dolven SI, Meyer A, Koehn RK, Rawson PD. 2000. Origin of the antitropical distribution pattern in marine mussels (Mytilus spp.): routes and timing of transequatorial migration. Mar Biol. 136:69–77.
- Mizi A, Zouros E, Moschonas N, Rodakis GC. 2005. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritance mode of mtDNA. Mol Biol Evol. 22:952–967.
- Oyarzún PA, Toro JE, Cañete JI, Gardner JPA. 2016. Bioinvasion threatens the genetic integrity of native diversity and a natural hybrid zone: smooth-shelled blue mussels (Mytilus spp.) in the Strait of Magellan. Biol J Linn Soc. 117:574–585.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Skibinski DO, Gallagher C, Beynon CM. 1994. Mitochondrial DNA inheritance. Nature. 368:817–818.
- Śmietanka B, Burzyński A, Wenne R. 2010. Comparative genomics of marine mussels (Mytilus spp.) gender associated mtDNA: rapidly evolving atp8. J Mol Evol. 71:385–400.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Venetis C, Theologidis I, Zouros E, Rodakis GC. 2007. A mitochondrial genome with a reversed transmission route in the Mediterranean mussel Mytilus galloprovincialis. Gene. 406:79–90.
- Westfall KM, Gardner JPA. 2010. Genetic diversity of southern hemisphere blue mussels (Bivalvia: Mytilidae) and the identification of non-indigenous taxa. Biol J Linn Soc. 101:898–909.
- Zbawicka M, Burzyński A, Wenne R. 2007. Complete sequences of mitochondrial genomes from the Baltic mussel Mytilus trossulus. Gene. 406:191–198.
- Zbawicka M, Wenne R, Burzyński A. 2014. Mitogenomics of recombinant mitochondrial genomes of Baltic Sea Mytilus mussels. Mol Genet Genomics. 289:1275–1287.
- Zouros E, Ball AO, Saavedra C, Freeman KR. 1994. Mitochondrial DNA inheritance. Nature. 368:818.
