Abstract
The complete mitochondrial genome of the edible fungus Thelephora ganbajun was determined using Illumina sequencing. This mitogenome is a circular molecule of 52,857 bp in length with a GC content of 25.73%. Gene prediction showed that the mitogenome codes 28 tRNAs, 2 pseudo-tRNAs, and 21 known and 7 hypothetical proteins. The evolutionary relationships between Th. ganbajun and other representative species based on the mitogenome are consistent with those based on nuclear genes. The mitogenome information of Th. ganbajun should contribute to our understanding of the diversity and evolution of Thelephorales.
Thelephora species are basidiomycetes and ectomycorrhizal (ECM) fungi (Tedersoo et al. Citation2014). Members of this genus are distributed globally, contributing significantly to plant health and ecosystem stability (Kõljalg Citation1995; Martini & Hentic Citation2005; Yorou & Agerer Citation2008). In Yunnan province of China, Thelephora ganbajun is among the best-known Thelephora species, distributes primarily in pine forests and has been heavily harvested as a food delicacy (Zang Citation1986, Citation1987; Zhang & Yang Citation2013). While the nuclear genetic diversity and bioactive compounds of Th. ganbajun have been investigated (Lin & Ji-Kai Citation2001; Sha et al. Citation2008; Yang et al. Citation2004), little is known about its mitochondrial genome. Here we report the complete mitogenome sequence of Th. ganbajun (KY245891) and provide a phylogenetic analysis of its relationships with several representative taxa based on concatenated mitochondrial protein-coding genes.
The mitogenome was extracted from the whole genome sequence of a pure culture of strain P2 (collected in Yunnan province) using the Illumina HiSeq-1TB platform. This strain has been deposited in State Key Laboratory for Conservation and Utilization of Bio-Resources at Yunnan University. To obtain the mitogenome, each fastq file was QC filtered and subsequently assembled using Velvet (Zerbino & Birney Citation2008). The resulting assembly was used to create a long mate-pair library with insert 3000 ± 300 bp which was further assembled with the original Illumina library using AllPathsLG (Gnerre et al. Citation2011), to produce a 176.8 × coverage main assembly containing 3 scaffolds. The gaps were filled by separate PCR and sequencing with primers on regions flanking the gaps, resulting in one circular mitochondrial genome. Annotation was performed using MFannot (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl) and the tRNAs and rRNAs were confirmed using RNAweasel (www.megasun.bch.umontreal.ca/RNAweasel/) and tRNAscan-SE (http://lowelab.ucsc.edu/tRNAscan-SE/) (Schattner et al. Citation2005). The general representation of the circular mitochondrial genome and the GC skew were prepared using the DNAPlotter software (Carver et al. Citation2009). The completely annotated mitogenome sequence is available in GenBank (accession KY245891).
The assembled mitochondrial genome was 52,857 bp in length with a GC content of 25.73%, coding for 28 tRNAs, 2 pseudo-tRNAs, 21 known proteins (including one pseudo gene and two overlapped polB2 genes), and 7 hypothetical proteins (or ORFs). The 28 tRNA genes covered 18 standard amino acids but without tRNA genes for Cysteine and Glutamic acid. Introns were found in three genes: nrDNA-LSU (2 group I A introns and 2 group I B introns), NAD5 (1 group I B intron) and COX1 (1 group I A intron and 1 group I B intron). Most introns have an ORF either of unknown function or code for a homing endonuclease.
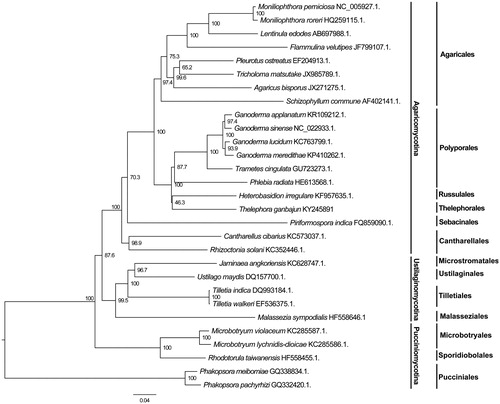
Our mitogenome is the first submitted Thelephorales mitogenome in the GenBank database. Based on the concatenated protein sequences, our analyses revealed that Th. ganbajun was a member of Agaricomycotina and closely related to Polyporales and Russulales (), consistent with the results obtained based on nuclear genes (Binder & Hibbett Citation2002; Garcia-Sandoval et al. Citation2011; Hibbett et al. Citation2007).
Figure 1. Phylogenetic analysis of 19 species of Agaricomycotina constructed using the Neighbour-Joining method as implemented in MEGA7.0 (Kumar et al. Citation2016) based on concatenated amino acid sequences of 14 mitochondrial protein-coding genes. The following 14 mitochondrial protein-coding genes were concatenated: atp6, atp8, atp9, cytb, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5 and nad6. The concatenated amino acid sequences were aligned using Clustal X (Thompson et al. Citation1997). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Binder M, Hibbett DS. 2002. Higher-level phylogenetic relationships of homobasidiomycetes (mushroom-forming fungi) inferred from four rDNA regions. Mol Phylogenet Evol. 22:76–90.
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 25:119–120.
- Garcia-Sandoval R, Wang Z, Binder M, Hibbett DS. 2011. Molecular phylogenetics of the Gloeophyllales and relative ages of clades of Agaricomycotina producing a brown rot. Mycologia. 103:510–524.
- Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 108:1513–1518.
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R. 2007. A higher-level phylogenetic classification of the Fungi. Mycol Res. 111:509–547.
- Kõljalg U. 1995. Tomentella (Basidiomycota) and related genera in temperate Eurasia. Synopsis Fungorum, Fungiflora, Oslo 9:1–213.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lin H, Ji-Kai L. 2001. Two novel phenylacetoxylated p-terphenyls from Thelephora ganbajun Zang. Z Naturforsch C J Biosci. 56:983–987.
- Martini E, Hentic R. 2005. Tomentella lilacinogrisea and T. guadelupensis sp. nov. Bull Soc Mycol Fr. 121:17–27.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Sha T, Xu J, Palanichamy MG, Zhang HB, Li T, Zhao ZW, Zhang YP. 2008. Genetic diversity of the endemic gourmet mushroom Thelephora ganbajun from south-western China. Microbiology. 154:3460–3468.
- Tedersoo L, Harend H, Buegger F, Pritsch K, Saar I, Kõljalg U. 2014. Stable isotope analysis, field observations and synthesis experiments suggest that Odontia is a non-mycorrhizal sister genus of Tomentella and Thelephora. Fungal Ecol. 11:80–90.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882.
- Yang WM, Liu JK, Hu L, Dong ZJ, Wu WL, Chen ZH. 2004. Antioxidant properties of natural p-terphenyl derivatives from the mushroom Thelephora ganbajun. Z Naturforsch C J Biosci. 59:359–362.
- Yorou NS, Agerer R. 2008. Tomentella africana, a new species from Benin (West Africa) identified by morphological and molecular data. Mycologia. 100:68–80.
- Zang M. 1986. Criticism on Thelephora ganbajun position. Edible Fungi. 4:1–2.
- Zang M. 1987. Some new and noteworthy higher fungi from eastern Himalayas. Acta Bot Yunnanica. 1:009.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang Q, Yang X. 2013. Situation, challenge and strategy of wild edible mushrooms export in Yunnan province. Technol Market. 20:316–318.
