Abstract
This study sequenced the entire mitochondrial genome of Populus davidiana Dode. It was 779,361 bp in length, containing 33 protein-coding genes, 3 rRNA genes and 22 tRNA genes and 1 pseudogene, and its GC content was 44.8%. Phylogenetic analysis was conducted using 6 mitochondrial genomes from the Salicaceae and Euphorbiaceae families, resulting that P. davidiana Dode was closely related to Populus tremula and Populus tremula × Populus alba. These results will provide fundamental data for the evolutionary studies in Populus genus.
Populus davidiana Dode is mainly distributed in the northeastern hemisphere. It belongs to the subsection trepidae of the section leuce, along with Populus tremula L. (European aspen), P. tremuloides Michx. (Quaking aspen) and P. grandiden (big-tooth aspen) (Lee et al. Citation2011). This species has been extensively planted in Korea for biomass production in short rotation plantations. In the genus populus, only two complete mitogenomes have been sequenced available for P. tremula (Genbank: KT337313) and Populus tremula × Populus alba (Genbank: KT429213) (Kersten et al. Citation2016). In this study, we provided the complete mitochondrial genome sequence (Genbank: KY216145) of P. davidiana Dode and compared its mitogenome sequences with those of six others available from the Salicaceae and Euphorbiaceae families.
The plant material of the female P. davidiana (Accession number, Odae 19) was collected from Kyungbuk-Youngju clonal plantation (36°49´N 128°31´E) in Republic of Korea, which was the same plant material previously used for the chloroplast genome sequencing (Choi et al. Citation2016). Total genomic DNA was extracted from fresh leaves using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA). The three independent sequencing libraries were prepared; shotgun library for 454 GS FLX and 3kb and size free mate-pair libraries for Illumina Hiseq 2500. A total of 1021M raw reads were retrieved and quality-trimmed with default parameters using SOAPec v2.03 (Beijing Genomics Institute, Shenzhen, China), Cutadapt (Department of Computer Science, TU Dortmund, Science for Life Laboratory), DeconSeq-standalone v0.4.3 and Sickle v1.33 (GitHub, Inc., San Francisco, CA). The resultant 935M trimmed reads were then used for the mitochondrial genome reconstruction using the PLATANUS assembler, by mapping them to the reference mitogenome of P. tremula. A total of 14,136,698 individual mitochondrial reads generated up to 2228 × average coverage. Subsequently the mitochondrial genome of P. davidiana was annotated using BLASTn and ORF Finder at NCBI by comparing with that of P. tremula.
The complete mitochondrial genome of P. densiflora is 779,361bp in length, which is smaller than P. tremula (783,442bp) and P. tremula × P. alba (783,513bp). The mitochondrial genome contains 59 genes, including 33 protein-coding genes, 3 ribosomal RNA genes, 22 transfer RNA genes and one pseudogene (rpl16). Among 33 protein-coding genes (a total of 57,067bp in length), the largest gene was nad4 (8370 bp) while atp9 was the smallest gene (224 bp). The GC content of the mitochondrial genome is 44.8%.
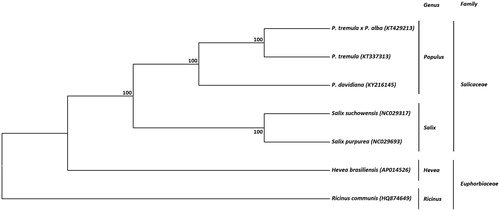
The complete mitochondrial genome of P. davidiana was aligned with those of 6 other species available from the Salicaceae and Euphorbiaceae families. Based on this alignment, a phylogenetic tree was generated by UPGMA method of MEGA 7 using 1000 bootstrap replicates (Kumar et al. Citation2016) (). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches (Felsenstein Citation1985). The evolutionary distances were computed by Maximum Composite Likelihood (MCL) method and are in the units of the number of base substitutions per site (Tamura et al. Citation2004). All Populous mtDNA sequences clustered together placing other three genera, Salix, Hevea, and Ricinus, as outgroups. The P. davidiana mtDNA sequence was very similar to both P. tremula and P. tremula × P. alba with the same nucleotide identity rate (99.0%).
Figure 1. Phylogenetic tree based on seven complete mitochondrial genome sequences from the Salicaceae and Euphorbiaceae families. The tree was constructed using UPGMA method and bootstrap support values (%) from 1000 replicates are shown above branches. GenBank accession numbers: Populus davidiana (KY216145), Populus tremula (KT337313), Populus tremula x Populus alba (KT429213), Salix suchowensis (NC029317), Salix purpurea (NC029693), Hevea brasiliensis (AP014526), Ricinus communis (HQ874649).
Disclosure statement
The authors report that they have no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Choi MN, Han M, Park HS, Kim MY, Kim JS, Na YJ, Sim SW, Park EJ. 2016. The complete chloroplast genome sequence of Populus davidiana Dode. Mitochondrial DNA B. 1:674–675.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791.
- Kersten B, Faivre Rampant P, Mader M, Le Paslier M-C, Bounon R, Berard A, Vettori C, Schroeder H, Leplé J-C, Fladung M. 2016. Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLoS ONE. 11:e0147209.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee KM, Kim YY, Hyun JO. 2011. Genetic variation in populations of Populus davidiana Dode based on microsatellite marker analysis. Genes Genomics. 33:163–171.
- Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 101:11030–11035.
