Abstract
In this study, the complete mitochondrial genome sequenced and analyzed from a fly, Muscina angustifrons which collected from South Korea. The size of mitochondrial genome is 16,316 bp with 40.9% A, 12.3% C, 8.4% G and 38.4% T distribution. Furthermore, phylogenetic relationships of the superfamily Muscoidea evaluated due to mitochondrial protein coding genes. The results showed that the family Muscidae is a paraphyletic group and the closest species to M. angustifrons is M. levida. This is the third complete mitochondrial genome for the genus Muscina and the first genus record from South Korea.
Muscina is a fly genus that belongs to the family Muscidae. They are represented with 27 species and abundant in worldwide (De Carvalho et al. Citation2005). Species of Muscina on the corpses are critically important for forensic investigation for determination of post-mortem intervals and estimation of the time of death (Byrd & Castner Citation2010). Therefore, correct identification of the insect species from crime scenes is crucial for forensic investigations. There are only two mitochondrial genome records from Muscina species in the GenBank which are M. stabulans (KM676394) from China and M. levida (KT272866) from U.S.A (Lan et al. Citation2015; Junqueira et al. Citation2016). In this study we are providing the complete mitochondrial genome of Muscina angustifrons. This is the third complete mitogenome belonging to the genus Muscina.
The specimens examined in the study have been collected from shade, forest, Mt. Gaewun, Seoul/37°35′43.0”N 129°01′43.1”E, August 2016. The collected specimens deposited in Department of Legal Medicine, Korea University (16An27). DNA preparation, DNA sequencing, mitochondrial gene annotation and phylogeny reconstruction methods described in our previous study (Karagozlu et al. Citation2016).
The mitochondrial genome of M. angustifrons (GenBank Accession number: KY495724) has typical gene order and gene orientation with ancestral insect genome (Cameron Citation2014). It consist of 13 protein-coding genes, two rRNA genes, 22 tRNA genes and one putative control region likewise the other Muscina records (Lan et al. Citation2015; Junqueira et al. Citation2016). Among them M. angustifrons has the longest mitochondrial genome with 16,316 bp length. The reason of this difference may be the length of the putative control region because it has the longest putative control region (1369 bp) which located between 12S rRNA gene and tRNA-Ile. Muscina angustifrons consists of seven overlapping regions in the genome with 1 to 40 bp lengths. The largest overlapping region is located between tRNA-Leu and 16S rRNA gene. The mitogenome shows 26 intergenic sequences varying from 1 to 110 bp in lengths. The longest intergenic sequence is between tRNA-Glu and tRNA-Phe.
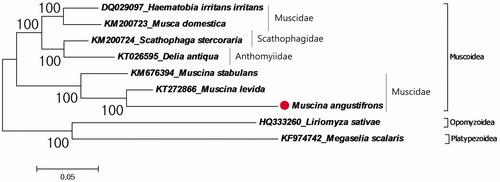
Furthermore phylogenetic relationship of M. angustifrons in the superfamily Muscoidea was investigated (). The phylogenetic tree showed that the closest species to M. angustifrons is M. levida in the family Muscidae. As representatives of the family Muscidae, M. angustifrons, M. levida and M. stabulans was clustered in a single group, while Haematobia irritans irritans and Musca domestica showed another group even though they are representative of the family. This suggests the family Muscidae is a paraphyletic group. Similar result showed by the previous mitochondrial and nuclear data based research (Dsouli et al. Citation2011). Although Muscidae flies are important insects for the forensic entomology (Benecke Citation2001), it is hard to identify specimens by morphological examination because of highly similar morphological appearance according to metamorphic stages (Lan et al. Citation2015). Therefore, complete mitochondrial genome sequences can be important tool in forensic entomology for identifying species.
Figure 1. Molecular phylogeny of M. angustifrons in the superfamily Muscoidea. The complete mitochondrial genomes for reconstruction of phylogenetic tree retrieved from GenBank and the records belonging to the superfamily Opomyzoidea and Platypezoidea chosen as representative of outgroup. M. angustifrons record marked with a dot.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIP) (No. NRF-2016M3A9E1915578) and Projects for Research and Development of Police science and Technology under Center for Research and Development of Police Science and Technology and Korean National Police Agency (No. PA-G000001-2016-101).
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Benecke M. 2001. A brief history of forensic entomology. Forensic Sci Int. 120:2–14.
- Byrd JH, Castner JL. 2010. Forensic entomology: the utility of arthropods in legal investigations. 2nd ed. Boca Raton: CRC Press; p. 705.
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39:400–411.
- De Carvalho CJB, Couri MS, Pont AC, Pamplona D, Lopes SM. 2005. A catalogue of the Muscidae (Diptera) of the Neotropical Region. Zootaxa. 860:1–282.
- Dsouli N, Delsuc F, Michaux J, Stordeur E, Couloux A, Veuille M, Duvallet G. 2011. Phylogenetic analyses of mitochondrial and nuclear data in haematophagous flies support the paraphyly of the genus Stomoxys (Diptera: Muscidae). Infect Genet Evol. 11:663–670.
- Junqueira ACM, Azeredo-Espin AML, Paulo DF, Marinho MAT, Tomsho LP, Drautz-Moses DI, Purbojati RW, Ratan A, Schuster SC. 2016. Large-scale mitogenomics enables insights into Schizophora (Diptera) radiation and population diversity. Sci Rep. 6:21762.
- Karagozlu MZ, Sung JM, Lee JH, Kwon T, Kim CB. 2016. Complete mitochondrial genome sequences and phylogenetic relationship of Elysia ornata (Swainson, 1840) (Mollusca, Gastropoda, Heterobranchia, Sacoglossa). Mitochondrial DNA B Resour. 1:230–232.
- Lan L, Liu Y, Yan J, Lin L, Zha L. 2015. The complete mitochondrial genome of the flesh fly, Muscina stabulans (Diptera: muscidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:4069–4070.
