Abstract
The hawksbill turtle, Eretmochelis imbricata (Linnaeus, 1766), is an endangered sea turtle in Colombian Caribbean beach. In this study, we report the complete mitochondrial DNA sequences of hawksbill turtle. The entire genome comprised 16,386 base pairs, and a nucleotide frequency of T: 25.6%, C: 26.9%, A: G 35.4% and 12.1%. The mitogenome sequence of hawksbill turtle would contribute to better understand population genetics, and evolution of sea turtles. Molecule was deposited at the GenBank database under the accession number KP221806.
The hawksbill turtle, Eretmochelys imbricata (Linnaeus, 1766), is a marine turtle belonging to the Cheloniidae family, order testudines. Hawksbill is a specie distributed throughout the tropical and central Atlantic and Indo-Pacific region (Lutz & Musick Citation1997). In Colombia nests on the coast the Pacific and Atlactic oceans. Hawksbill presents a way of life very complex and specialized. To mature, reach adulthood, reproduce and complete the life cycle, they need a variety of means, including terrestrial beaches, open sea, coastal and estuarine waters (Cuevas et al. Citation2008). Hawksbills reach maturity after 20–40 years (Bowen & Karl Citation2007). Actually, the hawksbill turtle is classified ‘Critically Endangered’ (CR) (Mortimer & Donnelly Citation2008), with increased risk of disappearing, for its low abundance, and by the continued looting of eggs (Castaño-Mora Citation2002; Daza-Criado & Hernández-Fernández Citation2014) and intense demand for the shields used in the manufacture of handicrafts (Chacón Citation2009). This illegal trade has led to a drastic decline in hawksbill populations as it is believed that the size of the world population has fallen by almost 80% over the last 100 years (Meylan Citation1999). For these reasons, the hawksbill turtle conservation is a priority at national and global level (Trujillo Citation2009). Phylogenetics, phylogeographics and conservation genetics analyses have been carried out using mitochondrial DNA to generate conscience of its conservation. Blood samples from an individual of hawksbill turtle from the Don Diego beach in Tayrona National Park, Santa Marta, Colombia, were collected following the Dutton (Citation1996) methodology. The sampled individuals are currently part of a head-starting project at the El Rodadero Aquarium and Museum (AMM El rodadero), an intermediate sector between Punta Gaira and Punta Cabeza de Negro (11°13"W, 74°14"N) in the city of Santa Marta, Department of Magdalena. The mtDNA of the hawksbill turtle was obtained by amplifying 24 DNA fragments of 800–1000 bp, and then were sanger sequenced. All sequences were assembled by means of the Geneious R6 program (Biomatters, Ltd., Auckland, New Zealand) and contigs carrying mitochondrial genes were identified against all BLASTX database. The phylogenetic tree was constructed used Geneious R6 and MEGA 5.2. (Tamura et al. Citation2011). The complete mitogenome of hawksbill turtle was annotated, which was 16,386 bp long, composed for 13 protein-coding genes (ND1, ND2, ND3, ND4L, ND4, ND5, ND6, COI, COII, COIII, ATP8, ATP6 and CytB), 22 tRNA, 2 rRNA (12S rRNA and 16S rRNA), and a non-coding control region (D-Loop) (). This mitogenome has a typical order of terrestrial, freshwater and marine turtles (Drosopoulou et al. Citation2012; Duchene et al. Citation2012) and vertebrates in general (Boore Citation1999). 15 tRNA, 2 rRNA, and 12 protein-coding genes are encoded by the H-strand, and 7 tRNA and 1 protein-coding gene (ND6) are encoded by the L-strand. Our analysis of the hawksbill mitogenome is the first report in the Colombian Caribbean region, generating basic information for future studies at a genetic level and contributing to management and conservation plans for this species.
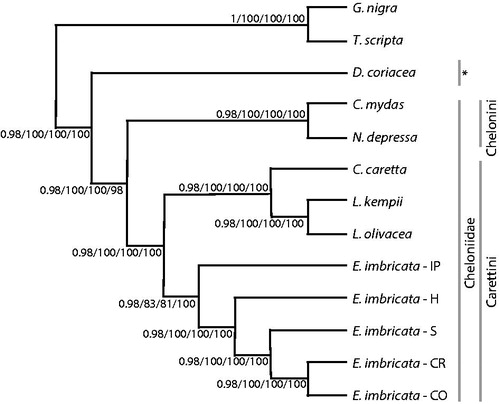
Figure 1. Philogenetics consensus by majority rule tree obtained using the algorithms neighbour-joining (NJ), maximum likelihood (ML), maximun parsimony (MP) and Bayesian inference (IB) used complete mitogenomes (17.384 pb). The topology of the tree shows correct association between turtles forming corresponding relationships between tribes and families. The values located in the nodes refer to the posterior probability of IB/and bootstrap, obtained with ML/MP/NJ methods. *Family Dermochelyidae.
Acknowledgements
We are grateful to Aquarium and Maritime Museum in Santa Marta Rodadero for collaboration in obtaining and providing samples of hawksbill turtles, Eretmochelys imbricata, for the development of this study. Samples were obtained under a research permit that was granted by the Ministry of Environment and Territorial Development (#24 of June 22, 2012) and Contract for Access to Genetic Resources (#64 of April 23, 2013).
Disclosure statement
The authors report no conflict of interest and also state that they are responsible for content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Bowen BW, Karl SA. 2007. Population genetics and phylogeography of sea turtles. Mol Ecol. 16:4886–4907.
- Castaño-Mora OV. 2002. Libro Rojo de Reptiles de Colombia. Bogotá: Instituto de Ciencias Naturales, Universidad Nacional de Colombia, Ministerio del Medio Ambiente, Conservación Internacional.
- Chacón D. 2009. Actualización del estado de la tortuga carey (Eretmochelys imbricata) en el Caribe y Atlántico occidental. Documento preparado para el Taller Regional sobre la Tortuga Carey en el Gran Caribe y Atlántico Occidental celebrado del 23 al 26 de septiembre de 2009; p. 120.
- Cuevas E, Abreu-Grobois FA, Guzmán-Hernández V, Liceaga-Correa MA, Van Dam RP. 2008. Post-nesting migratory movements of hawksbill turtles Eretmochelys imbricata in waters adjacent to the Yucatan Peninsula, Mexico. Endanger Species Res. 10:123–133.
- Daza-Criado L, J. Hernández-Fernández J. 2014. Molecular identification and first report of mitochondrial COI gene haplotypes in the hawksbill turtle Eretmochelys imbricata (Testudines: Cheloniidae) in the Colombian Caribbean nesting colonies. Genet Mol Res. 13:7123–7132.
- Drosopoulou E, Tsiamis G, Mavropoulou M, Vittas S, Katselidis KA, Schofield G, Palaiologou D, Sartsidis T, Bourtzis K, Pantis J, Scouras ZG. 2012. The complete mitochondrial genome of the loggerhead turtle Caretta caretta (Testudines: Cheloniidae): Genome description and phylogenetic considerations. Mitochondrial DNA. 23:1–12.
- Duchene S, Frey A, Alfaro-Núñez A, Dutton PH, Gilbert MTP, Morin PA. 2012. Marine turtle mitogenome phylogenetics and evolution. Mol Phylogenet Evol. 65:241–250.
- Dutton PH. 1996. Methods for collection and preservation of samples for Sea Turtle genetic studies. In: Proceedings of the International Symposium on Sea Turtle Conservation Genetics (Bowen BW and Witzell WN, eds.). NOAA Technical Memorandum, Miami; p. 17–24.
- Lutz PL, Musick JA. 1997. The biology of sea turtles. Boca Raton, Florida: CRC Press; p. 277–296.
- Meylan AB. 1999. Status of the hawksbill turtle (Eretmochelys imbricata) in the Caribbean region. Chelonian Conserv Biol. 3:177–184.
- Mortimer JA, Donnelly M, IUCN SSC Marine Turtle Specialist Group. 2008. Eretmochelys imbricata. The IUCN Red List of Threatened Species 2008: e.T8005A12881238. Available from: http://dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T8005A12881238.en.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Trujillo N. 2009. Caracterización genética de la tortuga carey (Eretmochelys imbricata) (Linneaus, 1766)) en Colombia, basada en la región control de ADNmt. Armenia: Universidad del Quindío.
