Abstract
We report the complete mitochondrial genome sequence of Cerion uva uva (Linnaeus 1758), the type species of the type genus of the family Cerionidae. The mitogenome is 15,043 bp in length, has a base composition of A (28.3%), T (34.4%), C (17.3%) and G (20.0%), and contains 13 protein-coding genes, 2 ribosomal RNA genes, as well as 22 transfer RNA genes. Gene order is the same as in Cerion incanum (Leidy 1851), but differs from those of all other Panpulmonata. This is the second mitochondrial genome sequenced within the family Cerionidae and will contribute to the assessment of the phylogeography of this family throughout the islands of the tropical western Atlantic.
Keywords:
Land snails of the Cerion uva complex are endemic to the islands of Aruba, Curaçao, and Bonaire (Baker Citation1924; De Vries Citation1974; Gould Citation1984). Cerion uva uva, the nominotypical subspecies of the type species of the type genus of the family Cerionidae, was isolated in eastern Curaçao during Pleistocene interglacial high sea-level stands (Wagenaar Hummelinck Citation1990), and was introduced to Aruba by humans within the past 800 years (Harasewych Citation2015). Recent phylogenetic analyses (Harasewych et al. Citation2011, Citation2015) have shown Cerion uva to be the most basal, and Cerion incanum (from the Florida Keys) to be among the most derived of the Cerionidae inhabiting the islands of the tropical western Atlantic.
We sequenced the complete mitochondrial genome of Cerion uva uva (GenBank accession number KY124261) using genomic DNA from a specimen (USNM 1153961L) collected at the type locality for this taxon (Schaarlo, Curaçao, 12° 6.42′ N, 68° 55.47′ W). Both partial COI and 16S sequences of this specimen have previously been published (Harasewych Citation2015). Extracted DNA was quantified using a Qubit dsDNA BR Assay Kit (ThermoFisher, Pittsburgh, PA). After quantification, DNA was sonicated (Qsonica Q800R2, QSonica, Newtown, CT) and libraries were prepared using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina® along with the NEBNext Multiplex Oligos for Illumina. Size selection of adaptor-ligated libraries (300–700 bp) was performed using the Pippin Prep. The Agilent TapeStation (Agilent, Santa Clara, CA) was used to validate library size selection. Libraries were quantified using qPCR (ViAA 7) to ensure generation of adaptor-ligated libraries. A 4nM library concentration was denatured for clonal amplification and sequencing on the Illumina Miseq, run at the Smithsonian’s Laboratories of Analytical Biology.
We obtained 4,808,211 sequences ranging in length from 35 to 301 bp. The mitogenome was assembled using the ‘map to reference’ feature of Geneious v. 9.1.6 (Biomatters, Newark, NJ) with the previously published (Harasewych Citation2015) partial COI sequence [Haplotype 57, GenBank KJ624976] from this specimen as the initial reference sequence. The mitogenome was independently reconstructed using the previously published partial 16S sequence [Haplotype 117, GenBank KJ636147] as the initial reference sequence. The two independently constructed mitogenomes were identical. A total of 10,398 reads mapped to the mitochondrial genome. Coverage ranged from 8× to 94× per site (46.5 ± 14.1). Mitochondrial elements were annotated using MITOS (Bernt et al. Citation2013), ARWEN (Laslett & Canbäck Citation2008) and the ORF finder in Geneious.
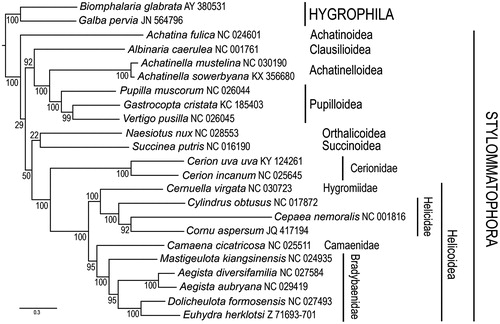
The mitochondrial genome of Cerion uva uva is a circular molecule consisting of 15,043 bp (134 bp shorter than C. incanum), with a base composition of A (28.3%), T (34.4%), C (17.3%) and G (20.0%). As in other pulmonates () (González et al. Citation2016), it contains 13 protein-coding genes, 2 ribosomal RNA genes, as well as 22 transfer RNA genes. Gene order is identical to that of Cerion incanum (González et al. Citation2016) (), but differs from that of other presently known Panpulmonata. This is the second mitochondrial genome sequenced within the family Cerionidae and will contribute to the assessment of the phylogeography of this family throughout the islands of the tropical western Atlantic.
Figure 1. Placement of Cerion uva uva among Stylommatophoran snails. Amino-acid sequences of protein-coding genes were individually aligned using the L-INS-i option with default parameters of the MAFFT v. 7 aligner (Katoh & Stanley Citation2013). Nucleotide alignments for individual protein-coding genes were obtained according to their amino-acid alignments using PAL2NAL (Suyama et al. Citation2006). Ribosomal genes were individually aligned using MAFFT (Q-INS-i option) (Katoh & Toh Citation2008). Individual nucleotide gene alignments were filtered using Gblocks (Talavera & Castresana Citation2007) with default parameters, allowing gaps in all positions. These were subsequently concatenated, leading to alignments with 9785 nucleotide positions for protein-coding genes and 1282 positions for the rRNA genes. Maximum likelihood analyses were performed using RAxML v. 8 (Stamatakis Citation2014). The general time reversible (GTR) model of nucleotide evolution was used. Maximum likelihood analyses consisted of 1000 independent tree searches and bootstrap runs. All analyses were run on the Smithsonian Institution’s high-performance computing cluster (SI/HPC). The resulting trees show similar relationships to previous studies (Gonzalez et al. Citation2016).
Acknowledgements
We are grateful to Gerard van Buurt for his guidance and assistance in collecting Cerion on Curaçao. This research was conducted, in part, while participating in the Smithsonian Institution's Deep Reef Observation Project (DROP) based at the Curaçao Sea Aquarium.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Baker HB. 1924. Land and freshwater molluscs of the Dutch Leeward Islands. Occasional papers of the Museum of Zoology, University of Michigan. Ann Arbor, MI, 152:1–160.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Stadler PF. 2013. MITOS: improved de novo Metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- De Vries W. 1974. Caribbean land molluscs: notes on Cerionidae. Stud Fauna Curaçao Caribbean Isl. 45:81–117, pls. 8?14.
- González VL, Kayal E, Halloran M, Shrestha Y, Harasewych MG. 2016. The complete mitochondrial genome of the land snail Cerion incanum (Gastropoda: Stylommatophora) and the phylogenetic relationships of Cerionidae within Panpulmonata. J Molluscan Stud. 82:525–533.
- Gould SJ. 1984. Covariance sets and ordered geographic variation in Cerion from Aruba, Bonaire and Curaçao: a way of studying non-adaptation. Syst Zool. 33:217–237.
- Harasewych MG. 2015. Systematics and phylogeography of Cerion sensu stricto (Mollusca: Gastropoda: Pulmonata: Cerionidae) from Aruba, Curaçao and Bonaire. J Molluscan Stud. 81:66–84.
- Harasewych MG, Sikaroodi M, Gillevet PM. 2011. The Delray Beach, Florida colony of Cerion (Paracerion) Tridentatum costellata Pilsbry, 1946 (Gastropoda: Pulmonata: Cerionidae): evidence for indirect Cuban origins. Nautilus. 125:159–163.
- Harasewych MG, Windsor AM, Lopez-Vera E, Thompson FG. 2015. On the phylogenetic relationships of the genus Mexistrophia and of the family Cerionidae (Gastropoda: Eupulmonata). Nautilus. 129:156–162.
- Katoh K, Stanley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9:286–298.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucelotide sequences. Bioinformatics. 24:172–175.
- Stamatakis A. 2014. RAxMl version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577.
- Wagenaar Hummelinck P. 1990. About the malacological subdivision of Curaçao; a review. Contr Zool. 60:181–187.
