Abstract
We sequenced the complete mitochondrial genome (mitogenome) for the North American Rhus gall aphid species Melaphis rhois. The mitogenome is 15,436 bp in length with a high A + T content of 82.7%, consisting of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. Its gene order is identical to that of the eastern Asian species Schlechtendalia chinensis. All protein-coding genes start with a typical ATN codon and terminate with a TAA codon except COI and ND4 by a single T residue. All the tRNAs except tRNACys formed a clover-leaf secondary structure. The mitogenome phylogeny of Aphididae suggests that M. rhois is most closely related to the eastern Asian Rhus gall aphid S. chinensis with the present sampling scheme.
The Rhus gall aphids belong to tribe Fordini (Hemiptera: Aphididae: Eriosomatinae) (Favret Citation2017) and were sometimes placed in subtribe Melaphidina (Remaudière & Remaudière Citation1997). Melaphidina aphids are primarily an Asian group with only one species Melaphis rhois in North America, exhibiting a biogeographic disjunction between eastern Asia and eastern North America (Wen Citation1999; Zhang et al. Citation1999; Ren et al. Citation2013). Up to now, only one complete mitochondrial genome (mitogenome) was reported in Eriosomatinae (Ren et al. Citation2016). We herein sequenced the complete mitochondrial genome of the unique North American M. rhois (GenBank accession KY624581).
All aphid individuals of a Rhus gall are of parthenogenetic generations. Melaphis rhois individuals from a gall were collected on Rhus glabra in Ohio (Columbus, 39°24′21.46″N, 84°23′37.20″W, altitude 238 m), USA (voucher deposited at the School of Life Science, Shanxi University, Taiyuan, China; Voucher no. A3037).
We obtained the mitogenome sequence of M. rhois using the shot-gun genome skimming method (Zimmer & Wen Citation2015) on an Illumina NextSeq 500 platform. The mitogenome sequence assembly used the eastern Asian species Schlechtendalia chinensis as the reference genome. We also did a de novo assembly with Velvet (Zerbino & Birney Citation2008).
The complete mitogenome of M. rhois is 15,436 bp in length, which contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA genes (rrnL and rrnS), and a control region. The gene order is the same as that of Schlechtendalia chinensis. All protein-coding genes have typical initiation and termination codons of ATN and TAA, respectively, except that COI and ND4 each terminate with a single T. The 22 tRNA genes range from 63 bp to 73 bp, and all except tRNACys exhibit a classical clover-leaf secondary structure, which we predicted with tRNAscan-SE v1.21 and/or RNAstructure (Lowe & Eddy Citation1997; Bellaousov et al. Citation2013). The tRNACys was determined based on comparisons to other aphid mitogenomes. The mitogenome of M. rhois has a base composition of A (44.6%), T (38.1%), C (11.3%), G (6.0%), and an A + T content (82.7%). The rrnL gene is 1270 bp with an A + T content 84.7%, while the rrnS gene is 772 bp with an A + T content 83.6%. The control region spans 703 bp and is located between rrnS and trnI with an A + T content 85.0%. The repeat region unique to some Aphidinae species is present in the close relative Schlechtendalia chinensis, but absent in M. rhois.
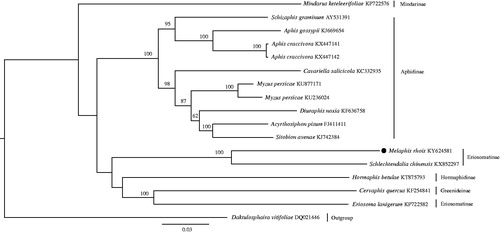
We employed the maximum-likelihood method to estimate the phylogeny of Aphididae using 13 PCG genes and 2 rRNA genes (). In this phylogeny, M. rhois is sister to Schlechtendalia chinensis. The mitogenome of M. rhois, the only North American species of the Rhus gall aphids, will be an important addition to explore the evolution of the biogeographic disjunction between eastern Asia and North America (Wen et al. Citation2016).
Figure 1. The maximum-likelihood tree of M. rhois and 16 accessions of Aphididae downloaded from GenBank using Daktulosphaira vitifoliae as an outgroup. Numbers above the branches indicate the bootstrap support values ≥50%. GenBank accession numbers and subfamily affiliations were indicated to the right of the terminals.
Acknowledgements
This study was partially supported by the National High Technology Research and Development "863" Program (2014AA021802), the National Natural Science Foundation of China (31170359), the Hundred-Talent Project in Shanxi Province, Shanxi Scholarship Council of China (2013-020), and the Laboratory of Analytical Biology of the National Museum of Natural History and the Endowment Grants Program of Smithsonian Institution. Computing was performed on the Smithsonian Institution High Performance Cluster (SI/HPC), Hydra.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Bellaousov S, Reuter JS, Seetin MG, Mathews DH. 2013. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 41:W471–W474.
- Favret C. 2017. Aphid Species File [Internet]. Version 5.0/5.0. [cited 2017 Feb 7]. Available from: http://Aphid.SpeciesFile.org
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Remaudière G, Remaudière M. 1997. Catalogue of the World’s Aphididae. Homoptera Aphidoidea. Paris, France: Institut National de la Recherche Agronomique.
- Ren ZM, Bai X, Harris AJ, Wen J. 2016. Complete mitochondrial genome of the Rhus gall aphid Schlechtendalia chinensis (Hemiptera: Aphididae: Eriosomatinae). Mitochond DNA B Res. 1:849–850.
- Ren ZM, Zhong Y, Kurosu U, Aoki S, Ma EB, von Dohlen CD, Wen J. 2013. Historical biogeography of eastern Asian–eastern North American disjunct Melaphidina aphids (Hemiptera: Aphididae: Eriosomatinae) on Rhus hosts (Anacardiaceae). Mol Phylogenet Evol. 69:1146–1158.
- Wen J. 1999. Evolution of the eastern Asia and eastern North America disjunct distributions in flowering plants. Ann Rev Ecol Syst. 30:421–455.
- Wen J, Nie ZL, Ickert-Bond SM. 2016. Intercontinental disjunctions between eastern Asia and western North America in vascular plants highlight the biogeographic importance of the Bering land bridge from late Cretaceous to Neogene. J Syst Evol. 54:469–490.
- Zhang GX, Qiao GX, Zhong TS, Zhang WY. 1999. Fauna Sinica Insecta, vol.14, Homoptera: Mindaridae and Pemphigidae. Beijing: Science Press.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zimmer EA, Wen J. 2015. Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches. J Syst Evol. 53:371–379.
