Abstract
Analysis of the marine black prickleback Xiphister atropurpureus Kittlitz using 76 bp paired-end Illumina sequences resulted in the assembly of its complete mitogenome. The mitogenome is 16,518 bp in length and contains an origin of light strand replication (OL), control region, 22 tRNA, 2 rRNA, and 13 protein-coding genes. Content and organization of the X. atropurpureus mitogenome is consistent with other teleost. Phylogenetic analysis of X. atropurpureus resolves it in a clade with another member of the Stichaeidae, Chirolophis japonicus Herzenstein.
The Stichaeidae fish family consists of entirely marine pricklebacks classified to six subfamilies, 35 genera, and 70 species (Eschmeyer & Fong Citation2017; Eschmeyer et al. Citation2017). Two mitogenomes have been deciphered for the family, these are Chirolophis japonicus subfamily Chirolophinae (Yang et al. Citation2016) and Leptoclinus maculatus Fries subfamily Lumpeninae (Swanburg et al. Citation2016). Here, we describe the mitogenome of X. atropurpureus, a member of the subfamily Xiphisterinae and common intertidal to subtidal species distributed from Kodiak Island, Alaska to Baja California, Mexico (Eschmeyer et al. Citation1983).
DNA was extracted from X. atropurpureus (Specimen Voucher- Hartnell College #262) collected from under a boulder in the mid intertidal at Pacific Grove, California (36°37′43.2″N, -121°55′17.5″W) following the protocol of Lindstrom et al. (Citation2011). The 76 bp paired-end library construction and sequencing was performed by myGenomics, LLC (Alpharetta, GA) yielding 16,427,262 reads. The mitogenome was assembled by mapping the reads against the reference sequence Chirolophis japonicus (GenBank NC_028022) using the Medium-Low Sensitivity/Fast setting in Geneious 8.0 (Biomatters Limited, Auckland, New Zealand). The genes were annotated using MITOS (Bernt et al. Citation2013) and adjusted manually using NCBI ORF-finder (https://www.ncbi.nlm.nih.gov/orffinder/). Alignment of the mitogenome to other perciforms was accomplished with MAFFT (Katoh & Standley Citation2013). The maximum-likelihood analysis was performed using complete mitogenome sequences with RaxML (Stamatakis Citation2014) in Galaxy (Giardine et al. Citation2005; Blankenberg et al. Citation2010; Goecks et al. Citation2010) using the GTR + gamma model and 1000 fast bootstraps. The tree was visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
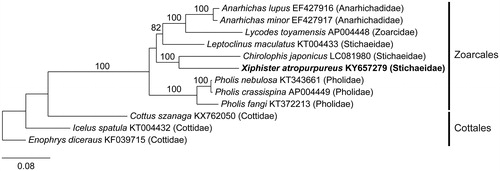
The mitogenome of X. atropurpureus (GenBank KY657279) is 16,518 bp in length, with a base composition of 25.4% A, 27.6% T, 18.7% G, and 28.3% C. It is highly conserved in organization, as is typical of animals (Boore Citation1999). The mitogenome contains 22 tRNA (trnL and trnS are duplicated), 2 rRNA (rrnL, rrnS), and 13 genes involved in electron transport and oxidative phosphorylation. All 13 genes start with the ATG initiation codon except cox1, which initiates with GTG. Most of the 13 genes terminate with the TAA stop codon, however, nd3 and cob terminate with TAG, and cox2 and nad4 with AGA. The nd6 gene and eight tRNAs encode on the light-strand, with the remaining genes encoding on the heavy strand. The OL is located between trnN and trnC, and is 38 bp in length, and the control region is 854 bp. Phylogenetic analysis of X. atropurpureus resolves it with two Stichaeids in a clade with C. japonicas, paraphyletic with respect to L. maculatus ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Ch. 10:11–21.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469.
- Eschmeyer WN, Fong JD. Species by family/subfamily [Internet]; [cited 2017 Feb 22]. Available from: http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp.
- Eschmeyer WN, Fricke R, van der Laan R. Catalog of fishes: genera, species, references [Internet]; [cited 2017 Feb 22]. Available from: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
- Eschmeyer WN, Herald ES, Hamman H. 1983. A field guide to Pacific coast fishes: North America. Boston (MA): Houghton Mifflin Company.
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15:1451–1455.
- Goecks J, Nekrutenko A, Taylor J, The Galaxy Team. 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lindstrom SC, Hughey JR, Martone PT. 2011. New, resurrected and redefined species of Mastocarpus (Phyllophoraceae, Rhodophyta) from the northeast Pacific. Phycologia. 50:661–683.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Swanburg T, Horne JB, Baillie S, King SD, McBride MC, Mackley MP, Paterson IG, Bradbury IR, Bentzen P. 2016. Complete mitochondrial genomes for Icelus spatula, Aspidophoroides olrikii and Leptoclinus maculatus: pan-Arctic marine fishes from Canadian waters. Mitochondr DNA Part A. 27:2982–2983.
- Yang H, Bao X, Wang B, Liu W. 2016. The complete mitochondrial genome of Chirolophis japonicus (Perciformes: Stichaeidae). Mitochondr DNA Part A. 27:4419–4420.