Abstract
After the establishment of an endosymbiotic relationship between a proto-mitochondrion and its probable archaeal host, mitochondrial genomes underwent a spectacular reductive evolution. An interesting pathway was chosen by mitogenomes of unicellular protists called dinoflagellates, which experienced an additional wave of reduction followed by amplification and rearrangement leading to their secondary complexity. The former resulted in a mitogenome consisting of only three protein-coding genes, the latter in their multiple copies being scattered across numerous chromosomes and the evolution of complex processes for their expression. These stunning features raise a question about the future of the dinoflagellate mitochondrial genome.
1. Introduction
Mitochondria are two-membrane-bounded cellular ‘powerhouses’ that evolved, very likely, as a result of syntrophy between a Rickettsia-like α-proteobacterium and a hydrogen-dependent archaeon around two billions years ago (Martin & Müller Citation1998; Martin et al. Citation2015; Wang & Wu Citation2015; Sousa et al. Citation2016). Their symbiosis triggered one of the most important transitions in the history of life, namely the transformation of prokaryotes into eukaryotes (Lane & Martin Citation2010).
During eukaryogenesis, the genome of the proto-mitochondrion underwent a tremendous reductive evolution, involving the loss of several thousand genes, either by being transferred to the host nuclear genome or by becoming irretrievably lost (Martin & Herrmann Citation1998; Timmis et al. Citation2004). Highly reduced mitochondria found in e.g. diplomonads, some amoebozoans, and microsporidians are called hydrogenosomes and mitosomes. The latter eliminated all their genetic material, representing an advanced level of reduction (Tachezy Citation2008; Hjort et al. Citation2010). The reductive evolution went to extremes in the oxymonad Monocercomonoides sp. as it lost the organelle itself (Karnkowska et al. Citation2016).
An interesting evolutionary pathway has been chosen by the mitochondrial genome (mitogenome) of dinoflagellates, which represents a remarkable trend towards simplicity and complexity at the same time. These unicellular protists play an important ecological role as ocean primary producers, parasites, and symbionts, e.g. of reef-building corals (Gómez Citation2012). They are distinguished by a number of peculiarities, among them the unique mitogenome and, in some cases, even two functional mitochondrial sets in one cell: one of their own and the second present in an engulfed endosymbiotic diatom (Imanian et al. Citation2012; Gagat et al. Citation2014). Together with their sister parasitic lineage Apicomplexa and some other relatives, dinoflagellates constitute the Myzozoa assemblage that unites with free-living ciliates in the superphylum Alveolata (; Burki Citation2014).
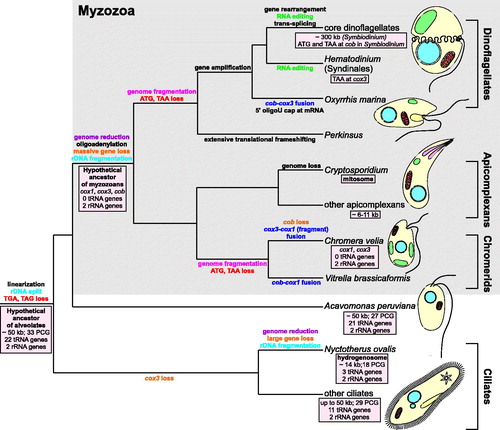
Figure 1. Evolution of mitochondrial genomes in alveolates. The ancestral genome of the superphylum Alveolata was probably about 50 kb in size, such as the mitogenomes of extant ciliates and Acavomonas peruviana, and encoded about 60 genes (Janouskovec et al. Citation2013). Since alveolates diverged about 850 million years ago (Berney & Pawlowski Citation2006), its mitogenome was subjected to various, often convergent, modifications and transformations as subsequent lineages of the superphylum radiated. Initially, the alveolate mitogenome was linearized from a circular form, lost two stop codons (TGA, TAG) and the genes coding for large and small ribosomal subunits were split into two separately encoded fragments. The most spectacular genome reduction occurred in the common ancestor of myzozoans, i.e. apicomplexans and dinoflagellates, and related lineages (e.g. chromerids), after their divergence from Acavomonas peruviana (Janouskovec et al. Citation2013). Extensive gene loss and gene transfer to the nuclear genome resulted in their extremely small mitogenomes containing only three protein-coding genes (cox1, cox3, and cob), sometimes fused, and two rRNA genes, which were subjected independently to further fragmentation in chromerids and the ancestor of dinoflagellates and perkinsids. This extremely small set of genes was even further reduced in Chromera velia as the chromerid completely lost the cob gene (Obornik & Lukes Citation2015). The myzozoan mitogenomes also got rid of about 20 tRNA genes present in the ancestral alveolate genome. At that time, oligoadenylation of transcripts probably evolved. A similar substantial genome reduction also occurred in the anaerobic ciliate Nyctotherus ovalis, whose mitochondrion was transformed into a hydrogenosome (de Graaf et al. Citation2011). This organelle produces hydrogen, which is utilized by methane-producing archaea living together with Nyctotherus as endosymbionts in the hindgut of cockroaches (de Graaf et al. Citation2011). The genome reduction went to extremes in the respiratory and intestinal parasite Cryptosporidium that completely lost its mitochondrial genetic material and transformed its mitochondrion into a mitosome, probably involved in Fe-S cluster assembly (Keithly et al. Citation2005). In dinoflagellates, genes were amplified to numerous copies, which resulted in an increase in their genome size amounting e.g. to ∼300 kb in Symbiodinium (Shoguchi et al. Citation2015). In these protists and sister lineages, various interesting molecular mechanism evolved, such as: translational frameshifting, the addition of 8–9 uridine caps at 5′ end of mRNAs, trans-splicing, and RNA editing (Flegontov & Lukeš Citation2012). The latter evolved, probably independently, in core dinoflagellates and Syndinales because editing sites are not conserved between these groups, which are separated by lineages without RNA editing (not shown in the figure; Flegontov & Lukeš Citation2012). It is assumed that universal start (ATG) and stop (TAA) codons, still present in apicomplexans, were independently lost in chromerids and the perkinsid-dinoflagellate branch. Given this model, the presence of TAA at the cox3 gene in Hematodinium (Jackson et al. Citation2012) as well as ATG and TAA at cob in Symbiodinium (Shoguchi et al. Citation2015) implies that these codons may have originated de novo. Alternatively, these codons might represent an ancestral state and many alveolates (Perkinsus, Oxyrrhis, and other dinoflagellates) lost these codons independently. PCG: protein-coding genes.
2. Dinoflagellate mitogenome content
The dinoflagellate mitochondrial genome has a similar gene content to the mitogenomes of other myzozoans, which are the most gene-impoverished mitochondrial genomes known (Waller & Jackson Citation2009; Flegontov & Lukeš Citation2012). They encode the same set of three divergent protein-coding genes: cob (cytochrome b), cox1 (cytochrome c oxidase subunit 1), and cox3 (cytochrome c oxidase subunit 3), two highly fragmented rRNA genes for large (LSU) and small (SSU) ribosomal subunits, as well as some uncharacterized small RNA fragments (Shoguchi et al. Citation2015). Compared to the mitogenome of their closest known Alveolata relative, Acavomonas peruviana, the myzozoan mitogenomes must have lost 45 genes (; Janouskovec et al. Citation2013). It is most likely the largest reduction discovered in any aerobic mitochondrion. At least 30 of the 45 genes, including tRNA and NADH dehydrogenase genes, were irretrievably lost. Therefore, myzozoan mitochondria must rely on imported, nucleus-encoded tRNAs to translate their proteins, and on an alternative type 2 NADH dehydrogenase to transfer electrons to ubiquinone (Flegontov et al. Citation2015). Nine of the 45 genes were transferred to the nuclear genome, e.g. cox2 (cytochrome c oxidase subunit 2), which is generally present in other eukaryotic mitogenomes. The other six genes, encoding ribosomal proteins, were either lost or their nuclear copies are too divergent to be recognized by computer algorithms (Janouskovec et al. Citation2013).
3. Dinoflagellate mitogenome structure
Although dinoflagellate mitochondrial genomes only have a few genes, they are anything but simple. Pulse field gel electrophoresis experiments indicate that they consist of multiple linear chromosomes with a size of 6–10 kb and longer (see Flegontov & Lukeš Citation2012 and references therein). The mitogenome size of Symbiodinium minutum, a reef-building coral endosymbiont, amounts to ∼326 kb, including mostly (99%) noncoding sequences. It is more than 50 times the size of the Plasmodium falciparum mitogenome (an apicomplexan that causes malaria), and 20 times the size of ours (Ji et al. Citation1996; Shoguchi et al. Citation2015). Still, the dinoflagellate mitochondrial genomes are smaller than the megabase-sized mitogenomes of some plants, e.g. in the genus Silene (Silene conica 11.3 Mb; Sloan et al. Citation2012).
Years of extensive studies have revealed that the large size of the dinoflagellate mitogenome is due to the numerous amplification and recombination events. They resulted in multiple copies of each gene and gene fragments linked in numerous configurations (Nash et al. Citation2008). The genes are often flanked by species-specific inverted repeats capable of forming stem-loop structures. Although the role of the inverted repeats is unknown, they might be involved, like in other organisms, in mitogenome recombination, replication, and transcript stability (reviewed by Flegontov & Lukeš Citation2012).
4. More peculiarities: missing ORFs boundaries, RNA editing, trans-splicing, and gene fusions
Very few genes and such immense genomes make a unique duet, but there are more peculiarities hidden in the dinoflagellate mitogenomes. First of all, it is difficult to find where the ORFs’ (Open Reading Frames) boundaries are, as the canonical start and stop codons were reported to be missing from the dinoflagellate transcripts (reviewed by Flegontov & Lukeš Citation2012). However, similar to ciliates and apicomplexans, S. minutum does contain unconventional start codons (AUU for Ile, AUA for Ile), and all investigated dinoflagellates use polyadenylation to generate a classical stop codon from an incomplete one ending with uracile in cox3, a phenomenon also observed in mitogenomes of some vertebrates. Poly(A) stretches might also constitute a dinoflagellate translation termination signal, e.g. in cox1, by causing a kind of ‘sliding’ movement of the ribosome that prematurely terminates translation (Koutmou et al. Citation2015). Only the cob gene, in S. minutum at least, has both canonical start and stop codons (Shoguchi et al. Citation2015). This means that stop codons are still used in dinoflagellates and have not been, for example, reassigned to other amino acids.
The transcripts of all protein-coding genes in dinoflagellates, and some rRNA fragments as well, are edited. The editing intensity increases from basal phylogenetic lineages, where it is absent, e.g. in Oxyrrhis marina, to the later branching ones known as the core dinoflagellates (; Zhang et al. Citation2008). Although RNA editing occurs in other organisms, the versatility and scale of changes is unprecedented in dinoflagellates. Nearly all possible substitutions (9 of 12) have been observed and they concern up to 6% of the nucleotides in dinoflagellate transcripts (Waller & Jackson Citation2009). Generally, editing increases GC content, thereby facilitating the use of nucleus-encoded tRNA. The changes may also eliminate incompatibilities between nucleus- and mitochondrion-encoded proteins, for example, by restoring evolutionarily conserved amino acids (Greiner & Bock Citation2013). The editing also get rid of an in-frame stop codons, e.g. in cox1 of Amphidinium carterae (Waller & Jackson Citation2009). Interestingly, beside land plants, dinoflagellates are the only group, for which plastid RNA editing has been reported, however, the process is not as widespread and elaborate as in their mitochondria (Knoop Citation2011; Smith & Keeling Citation2015).
The other ‘peculiarity’ concerns cox3 trans-splicing that is characteristic of all investigated core dinoflagellates (; Jackson & Waller Citation2013). The gene is broken between the regions coding for the sixth and seventh transmembrane helices. Therefore, in order to create the mature cox3 mRNA, the precursor transcripts must be joined. This process is imperfect because, depending on the species, a certain number of adenosine nucleotides are added between the two mRNAs, e.g. five in Karlodinium veneficum and ten in A. carterae, which in turn results in one or more lysines in the Cox3 protein. This is, however, tolerated due to the location of the hydrophilic insertion between the transmemebrane domains (Jackson & Waller Citation2013).
Contrary to split of cox3 in core dinoflagellates, independent mitochondrial gene fusion has been reported in the early-branching dinoflagellate Oxyrrhis marina (cob-cox3) and the chromerid Vitrella brassicaformis (cob-cox1) (Slamovits et al. Citation2007; Obornik & Lukes Citation2015). In Chromera veila, a putative cox3 was also fused with an upstream fragment of cox1. As the genes encode subunits of different electron transport complexes, they must function as separate proteins, and therefore they should be (i) translated individually from a polycistronic transcript, (ii) translated from separated transcripts produced by cleavage of pre-mRNA, or (iii) cleaved after their translation. It is also possible that Oxyrrhis and chromerids evolved different mechanisms to manage the fused genes and their products.
5. Is the dinoflagellate mitogenome going to be lost?
Such a small number of protein-coding genes raises a question about the future of the mitochondrial DNA in dinoflagellates, and other myzozoans as well, especially taking into account the fact that some apicomplexans (Cryptosporidium spp.) indeed have highly morphologically and functionally reduced mitochondria without DNA, i.e. mitosomes (Liu et al. Citation2016). But mitosomes, hydrogenosomes and other mitochondrion-related organelles (reviewed by Makiuchi & Nozaki Citation2014) have only evolved in parasitic lineages under anaerobic/hypoxic conditions, in which the energetic resources of the host are aplenty. In such environments, the abundance of host metabolites affects not only the mitochondrial genome but also the nuclear and plastid genomes as well. Cryptosporidium spp. perfectly exemplifies this through its loss of mitochondrial DNA, the plastid itself, and the great reduction of its nuclear genome in comparison to other eukaryotes, including apicomplexans (reviewed by Liu et al. Citation2016).
In contrast to Cryptosporidium spp., dinoflagellates live in changing, low-nutrient environments that favour highly adaptive and innovative species. In such conditions, organisms are especially under pressure to outcompete rivals, e.g. by growing faster, offering one of reasons why many dinoflagellates are mixotrophs, i.e. can both photosynthesize and feed by phagocytosis (Hansen Citation2011). The mitogenome loss in dinoflagellates seems highly incomprehensible because it would mean abandoning the most effective way of releasing energy from nutrients, i.e. oxidative phosphorylation. According to the CoRR (Co-location for Redox Regulation) hypothesis (Allen Citation2015; Allen & Martin Citation2016), at least one component of each existent respiratory chain complex (complex III and IV in dinoflagellates) must be encoded by the mitogenome to ensure its fine-tuned regulation, and consequently regulation of the entire organelle and cellular metabolism.
It cannot be ruled out that the ancestors of extant myzozoans once led parasitic lives. As a result of a relaxed selection, their mitogenomes were reduced and peculiarities such as, e.g. the missing start/stop codons and gene fragmentation appeared. When dinoflagellates diverged from apicomplexans and changed their trophic strategies, the increased energetic pressure could have triggered their mitogenome inflation. The genome expansion can increase the number of gene copies and consequently their products involved in oxidative phosphorylation, resulting in boosting energy production. Numerous copies also secure mitochondrial genes from accumulating deleterious mutations in an irreversible manner according to Muller’s ratchet (Martin & Herrmann Citation1998). The other complex processes necessary to express the remaining mitochondrial genes, such as RNA editing, may have evolved, to correct mutations introduced into these genes or increase diversity of their products, like in trypanosomes (Ochsenreiter & Hajduk Citation2006; Ochsenreiter et al. Citation2008). All the peculiarities in dinoflagellate mitochondrial genomes observed today result from the reductive forces that have been driving organellar evolution from the very beginning of eukaryogenesis.
Acknowledgements
We are grateful to Michael Nikiel for his language editing.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Allen JF. 2015. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc Natl Acad Sci USA. 112:10231–10238.
- Allen JF, Martin WF. 2016. Why have organelles retained genomes? Cell Syst. 2:70–72.
- Berney C, Pawlowski J. 2006. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc Biol Sci. 273:1867–1872.
- Burki F. 2014. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol. 6:a016147.
- de Graaf RM, Ricard G, van Alen TA, Duarte I, Dutilh BE, Burgtorf C, Kuiper JW, van der Staay GW, Tielens AG, Huynen MA, et al. 2011. The organellar genome and metabolic potential of the hydrogen-producing mitochondrion of Nyctotherus ovalis. Mol Biol Evol. 28:2379–2391.
- Flegontov P, Lukeš J. 2012. Mitochondrial genomes of photosynthetic euglenids and alveolates. In: Advances in Botanical Research. Oxford: Elsevier Ltd Academic Press; p. 127–153.
- Flegontov P, Michalek J, Janouskovec J, Lai DH, Jirku M, Hajduskova E, Tomcala A, Otto TD, Keeling PJ, Pain A, et al. 2015. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol Biol Evol. 32:1115–1131.
- Gagat P, Bodył A, Mackiewicz P, Stiller J. 2014. Tertiary plastid endosymbioses in Dinoflagellates. In: Endosymbiosis. Vienna: Springer; p. 233–290.
- Gómez F. 2012. A quantitative review of the lifestyle, habitat and trophic diversity of dinoflagellates (Dinoflagellata, Alveolata). Syst Biodivers. 10:267–275.
- Greiner S, Bock R. 2013. Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays. 35:354–365.
- Hansen PJ. 2011. The role of photosynthesis and food uptake for the growth of marine mixotrophic dinoflagellates. J Eukaryot Microbiol. 58:203–214.
- Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. 2010. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci. 365:713–727.
- Imanian B, Pombert JF, Dorrell RG, Burki F, Keeling PJ. 2012. Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts. PLoS One. 7:e43763.
- Jackson CJ, Gornik SG, Waller RF. 2012. The mitochondrial genome and transcriptome of the basal dinoflagellate Hematodinium sp.: character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biol Evol. 4:59–72.
- Jackson CJ, Waller RF. 2013. A widespread and unusual RNA trans-splicing type in dinoflagellate mitochondria. PLoS One. 8:e56777.
- Janouskovec J, Tikhonenkov DV, Mikhailov KV, Simdyanov TG, Aleoshin VV, Mylnikov AP, Keeling PJ. 2013. Colponemids represent multiple ancient alveolate lineages. Curr Biol. 23:2546–2552.
- Ji YE, Mericle BL, Rehkopf DH, Anderson JD, Feagin JE. 1996. The Plasmodium falciparum 6 kb element is polycistronically transcribed. Mol Biochem Parasitol. 81:211–223.
- Karnkowska A, Vacek V, Zubáčová Z, Treitli SC, Petrželková R, Eme L, Novák L, Žárský V, Barlow LD, Herman EK, et al. 2016. A eukaryote without a mitochondrial organelle. Curr Biol. 26:1274–1284.
- Keithly JS, Langreth SG, Buttle KF, Mannella CA. 2005. Electron tomographic and ultrastructural analysis of the Cryptosporidium parvum relict mitochondrion, its associated membranes, and organelles. J Eukaryot Microbiol. 52:132–140.
- Knoop V. 2011. When you can't trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci. 68:567–586.
- Koutmou KS, Schuller AP, Brunelle JL, Radhakrishnan A, Djuranovic S, Green R. 2015. Ribosomes slide on lysine-encoding homopolymeric A stretches. eLife. 4:e05534.
- Lane N, Martin W. 2010. The energetics of genome complexity. Nature. 467:929–934.
- Liu S, Roellig DM, Guo Y, Li N, Frace MA, Tang K, Zhang L, Feng Y, Xiao L. 2016. Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genomics. 17:1006.
- Makiuchi T, Nozaki T. 2014. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie. 100:3–17.
- Martin W, Herrmann RG. 1998. Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol. 118:9–17.
- Martin W, Müller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature. 392:37–41.
- Martin WF, Garg S, Zimorski V. 2015. Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond, B, Biol Sci. 370:20140330.
- Nash EA, Nisbet RE, Barbrook AC, Howe CJ. 2008. Dinoflagellates: a mitochondrial genome all at sea. Trends Genet. 24:328–335.
- Obornik M, Lukes J. 2015. The organellar genomes of Chromera and Vitrella, the phototrophic relatives of apicomplexan parasites. Annu Rev Microbiol. 69:129–144.
- Ochsenreiter T, Cipriano M, Hajduk SL. 2008. Alternative mRNA editing in trypanosomes is extensive and may contribute to mitochondrial protein diversity. PLoS One. 3:e1566.
- Ochsenreiter T, Hajduk SL. 2006. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep. 7:1128–1133.
- Shoguchi E, Shinzato C, Hisata K, Satoh N, Mungpakdee S. 2015. The large mitochondrial genome of Symbiodinium minutum reveals conserved noncoding sequences between dinoflagellates and apicomplexans. Genome Biol Evol. 7:2237–2244.
- Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. 2007. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol. 372:356–368.
- Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR. 2012. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 10:e1001241.
- Smith DR, Keeling PJ. 2015. Mitochondrial and plastid genome architecture: reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 112:10177–10184.
- Sousa FL, Neukirchen S, Allen JF, Lane N, Martin WF. 2016. Lokiarchaeon is hydrogen dependent. Nature Microbiology. 1:16034.
- Tachezy J. 2008. Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Berlin, Heidelberg: Springer.
- Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 5:123–135.
- Waller RF, Jackson CJ. 2009. Dinoflagellate mitochondrial genomes: stretching the rules of molecular biology. BioEssays. 31:237–245.
- Wang Z, Wu M. 2015. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep. 5:7949.
- Zhang H, Bhattacharya D, Maranda L, Lin S. 2008. Mitochondrial cob and cox1 genes and editing of the corresponding mRNAs in Dinophysis acuminata from Narragansett Bay, with special reference to the phylogenetic position of the genus Dinophysis. Appl Environ Microbiol. 74:1546–1554.
