Abstract
Glyptothorax cavia, a small-sized benthic fish, distributed in southwest China. In the present study, the complete mitochondrial genome of G. cavia was sequenced to be 16,529 bp in length, including 13 protein-coding genes, 2 ribosomal RNAs, 22 transfer RNAs, a control region, and the origin of the light strand replication. The overall nucleotide composition was 31.15% A, 25.86% T, 27.64% C, and 15.35% G, with an A + T bias of 57.01%. The gene composition and the structural arrangement of the G. cavia complete mtDNA were identical to most of the other vertebrates. This will provide a useful tool for understanding the genetic diversity, population structure, and conservation status of G. cavia in the future.
Introduction
Mitochondrial DNA (mtDNA) gene order was proposed to be quite conserved within vertebrates based on the gene order of the initial genome sequence (Anderson et al. Citation1981; Bibb et al. Citation1981). Glyptothorax cavia (Siluriformes: Sisoridae) is an endemic fish species which mainly distributes in the Irrawaddy, Nujiang River and their tributaries in China. In recent years, the natural resource of this species has seriously declined, as a result of overharvesting, water contamination, and especially dam construction (Shao et al. Citation2016). In the long run, a good understanding of the genetic diversity and population structure of G. cavia is required in order to establish adequate management plans for the conservation of this species. To address these topics, we determined the complete mitochondrial genome sequence of G. cavia for the first time.
Specimens of G. cavia were collected from Nujiang River (25°50′0.34″N. 98°51′33.76″E) in March 2015. The caudal fins of 12 specimens were cut-off and preserved in 95% ethanol, then stored under −80 °C until DNA extraction. Total genomic DNA was isolated from the caudal fin by proteinase K digestion followed by the standard phenol/chloroform method (Sambrook & Russell Citation2001) and visualized on 1.5% agarose gel. 20 sets of primers were designed for PCR amplification on the basis of aligned mitogenome sequences of Glyptothorax trilineatus with the Accession NC_021608.1. In order to avoid errors of assembly, the complete mtDNA genome was aligned and checked with three reported mtDNA genome sequences of Sisoridae species G. zainaensis (Accession NC_029709.1); G. fokiensis (Accession NC_018769.1); G. trilineatus (Accession NC_021608.1). The assembled sequence was analyzed using the software MitoAnnotator (Iwasaki et al. Citation2013) and nucleotide composition was calculated by MEGA6 (Tamura et al. Citation2013).
The complete mtDNA sequence of G. cavia reported here has been deposited in GenBank under the accession number KY230517. The mitochondrial genome of G. cavia is a circular molecule of 16,529 nucleotides, which is similar to other vertebrates, including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a non-coding control region (D-loop). The overall nucleotide composition is 31.15%, 25.86%, 27.64%, and 15.35% for A, T, C, and G, with an A + T content of 57.01%, respectively. Except for a single protein-coding gene (ND6) and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer (UCN), tRNAGlu, and tRNAPr°) encoded on the L-strand. All the other genes were encoded on the H-strand. The first non-coding region is 885bp between tRNAPr° and tRNAPhe, and the second one is the origin of light-strand replication, which extends up to 31bp. It is located in a cluster of five tRNA genes (the WANCY region) between tRNAAsn and tRNACys gene.
Furthermore, the termination codon varies with TAA, TA–, T––, or TAG. Virtually, all of the 13 protein-coding genes show the regular initiation codon ATG with the sole exception of COI which started with GTG. Six protein-coding genes terminated with the complete stop codon TAA (ND1, COI, ATPase8, ND4L, and ND5) or TAG (ND6), while the rest ended with incomplete stop codon T–– (ND2, COII, COIII, ND3, ND4, and Cytb) or TA– (ATPase 6), which is quite typical among mtDNA genes in other fishes (Zhou et al. Citation2012; Wang et al. Citation2013).
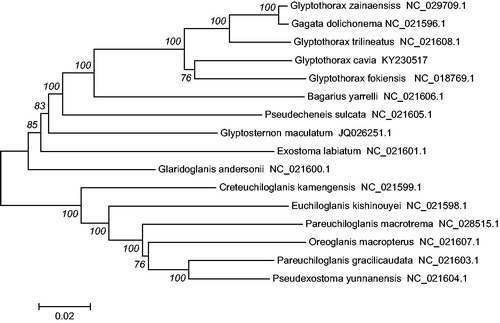
In addition, the mtDNA sequences of 15 species of fishes were downloaded from Gen Bank, Oreoglanis macropterus (Accession NC_021607.1), Pseudexostoma yunnanensis (Accession NC_021604.1), Pareuchiloglanis gracilicaudata (Accession NC_021603.1), P. macrotrema (Accession NC_028515.1) were used as an outgroup for phylogenetic analysis. Phylogenetic analyses were performed using the neighbour joining (NJ) in MEGA 6.0 (Kumar et al. Citation2008). The tree topologies based on complete mt DNA sequences in this study were identical and were statistically supported by high bootstrap and posterior probability values (). The mitogenome data provided strong support that G. Cavia was clustered together with Glyptothorax fokiensis (Accession NC_018769.1), G. trilineatus (Accession NC_021608.1), Gagata dolichonema (Accession NC_021596.1) and G. zainaensiss (Accession NC_029709.1). The phylogenetic analyses yielded convincing evidence that the genera Gagata and Glyptothorax constituted a monophyletic group and then formed a sister group with all Bagarius yarrelli (Accession NC_021606.1).
Figure 1. The consensus phylogenetic relationship of the G. cavia with other Sisoridae species. O. macropterus, P. yunnanensis, P. gracilicaudata, and P. macrotrema were used as an outgroup. The numbers along the branches are Bayesian posterior probability and bootstrap values for NJ, estimated for concatenated mitochondrial protein sequences.
Disclosure statement
The author is grateful to thank the open project funding (2015) of the Hubei Key Laboratory of Three Gorges Project for Conservation of Fishes (Grant No. 0704112). The author report no conflicts of interest. The author alone is responsible for the content and authorship of this report.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, De Bruijn M, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sange F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. 1981. Sequence and gene organization of mouse mitochondrial DNA. Cell. 26:167–180.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformatics. 9:299–306.
- Sambrook J, Russell D. Molecular cloning — a laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press, 2001.
- Shao K, Yan SX, Zhu B, Xu N, Li WT, Xiong MH. 2016. Complete mitochondrial genome of Pareuchiloglanis sinensis (Siluriformes: Sisoridae). Mitochondrial DNA. 27:713–714.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0 . Mol Biol Evol. 30:2725–2729.
- Wang XY, Cao L, Liang HW, Li Z, Zou GW. 2013. Mitochondrial genome of the Shorthead catfish (Pelteobagrus eupogon). Mitochondrial DNA. 24:1–2.
- Zhou C, Wang D, Yu M, He S. 2012. The complete mitochondrial genome of Glyptothorax fukiensis fukiensis (Teleostei, Siluriformes: Sisoridae). Mitochondrial DNA. 23:414–416.
