Abstract
Brachysomophis crocodilinus belongs to the family Ophichthidae, the complete mitochondrial genome of which was sequenced in this study. The mitochondrial genome of B. crocodilinus is of 17,818 bp in length, with overall base composition of 32.11% A, 24.69% T, 16.22% G, and 26.98% C. The genome content includes 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 2 control regions. The result of phylogenetic analysis indicates that B. crocodilinus mitogenome is close to that of Ophisurus macrorhynchos, which are nested within the family Ophichthidae.
Ophichthidae is a family of the order Anguilliformes, containing 59 genera and about 319 species. The distinguishing features of the family Ophichthidae are posterior nostril usually within or piercing upper lip; median supraorbital pore in frontal sensory canal; caudal fin absent; pectoral fins present or absent; vertebrae 110–270 (Nelson et al. Citation2016). Brachysomophis crocodilinus belongs to the family Ophichthidae, which distributes in the Indo-West Pacific; including East and South China Sea (Zhang Citation2010). In this study, the complete mitochondrial genome of B. crocodilinus was determined. Fish sample was collected from the Changjiang Estuary (30°51′16.24″N, 121°54′46.51″E), Shanghai, China. The specimen was stored in the Laboratory of Ichthyology, Shanghai Ocean University, with accession number 20160613. The sample DNA is available upon request.
Mitochondrial DNA is a maternally inherited circular genome that serves important functions in metabolism and population genetics (Boore Citation1999). In the present study, the sequenced mitochondrial genome of B. crocodilinus (GenBank accession number: KY081398) is of 17,818 bp in length, with overall base composition of 32.11% A, 24.69% T, 16.22% G, and 26.98% C. With the exception of two control regions, the genome content of B. crocodilinus includes 2 rRNA, 22 tRNA, and 13 protein-coding genes, as found in other vertebrates (Inoue et al. Citation2004). The mitogenome exhibits the translocation of nad6 in some species of the order Anguilliformes (Shen et al. Citation2014). The phenomenon also occurs in B. crocodilinus, arrangement of protein-coding genes of which is: nad1—nad2—cox1—cox2—atp8—atp6—cox3—nad3—nad4L—nad4—nad5—cob—nad6. Apart from the ND6 gene and eight tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser, Glu and Pro) encoded on the L-strand, most genes are on the H-strand. 11 of 13 protein-coding genes start with a common initiation codon ATG, while COI and ATP6 utilize GTG. According to the result given by Mitoannotator (Iwasaki et al. Citation2013), there are six protein-coding genes with the order of ND2, COII, ATP6, COIII, ND3, and ND4 ending with incomplete stop codon (T––, T––, TA–, TA–, T––, T––), while six genes (ND1, COI, ATP8, ND4L, ND5, Cytb) use the stop codon TAA, and ND6 uses the stop codon TAG. The end of ATP8 overlaps with the beginning of ATP6 with a length of 10 bp, and there is another overlap between ND4L and ND4 with a length of 7 bp.
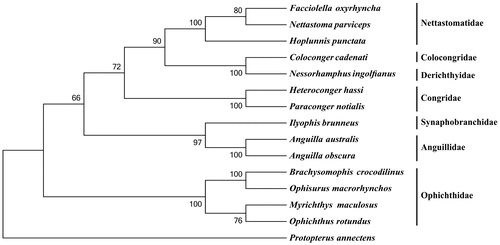
To investigate the phylogenetic relationship among the order Anguilliformes, we downloaded the mitochondrial genome sequences of 13 currently available species of Anguilliformes, including Anguilla australis (AP007235), A. obscura (AP007247), Coloconger cadenati (AP010863), Facciolella oxyrhyncha (AP010866), Heteroconger hassi (AP010859), Hoplunnis punctata (AP010865), Ilyophis brunneus (AP010848), Myrichthys maculosus (AP010862), Nessorhamphus ingolfianus (AP010850), Nettastoma parviceps (AP010864), Ophichthus rotundus (KY081397), Ophisurus macrorhynchos (AP002978) and Paraconger notialis (AP010860), together with African lungfish Protopterus annectens (NC018822) as an outgroup species. The phylogenetic tree was constructed using MEGA6 (Tamura et al. Citation2013) for neighbour-joining method. Tree topology was evaluated by 1000 bootstrap replicates. The result indicates that B. crocodilinus mitogenome is close to that of O. macrorhynchos, which are nested within the family Ophichthidae ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Inoue JG, Miya M, Tsukamoto K, Nishida M. 2004. Mitogenomic evidence for the monophyly of elopomorph fishes (Teleostei) and the evolutionary origin of the leptocephalus larva. Mol Phylogenet Evol. 32:274–286.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Mabuchi K, Takeshima H, Miya M. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world. 5th ed. New York: John Wiley and Sons, Inc.
- Shen X, Tian M, Meng XP, Cheng HL, Yan BL. 2014. Eels mitochondrial protein-coding genes translocation and phylogenetic relationship analyses. Acta Oceanol Sin. 36:73–81.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhang 2010. Fauna Sinica, Osteichthyes, Anguilliformes & Notacanthiformes. Beijing: Science Press.