Abstract
In this study, the complete mitochondrial genome of Carassioides acuminatus was first sequenced and annotated. The entire mitogenome is 16,579bp in length, which consists of 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region (D-loop). The overall nucleotide composition of the C. acuminatus mitochondrial genome shows an obvious anti-G bias. The accuracy of the fresh sequences was verified by phylogenetic analysis. The complete mitochondrial genome of C. acuminatus is useful to population genetics and molecular systematics.
Carassioides acuminatus (C. acuminatus) is a small-to moderate-sized freshwater economic fish belonging to the family Cyprinidae, order Cypriniformes (Zheng Citation1989). It is mainly distributed in the Pearl River and Hainan Island water system in China. As the C. acuminatus geographical distribution range is narrow, few studies have been reported (Chunxing et al. Citation2014). Due to its haploid nature, limited recombination, maternal inheritance, and rapid evolutionary rate, the mitochondrial DNA has now been widely used for studying population genetics, phylogeography and phylogeny and species identification. Consequently, it was necessary to determine the complete mitochondrial genome of the C. acuminatus and identify its phylogenetic relationships with the closely related species for the sustainable utilization of the C. acuminatus fishery resource.
In this study, the sample of the C. acuminatus was obtained from Lingao section (19°34′–20°02′ N, 109°3′–109°53′ E) in the Wenlan River, Hainan, China. It was stored in the Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China with the sample number CAWL2014. The complete mitochondrial genome of the C. acuminatus was sequenced using Illumina-based de novo transcriptome technology and annotated using bioinformatic tools (Laslett & Canbäck Citation2008; Tamura et al. Citation2013). The mitochondrial genome of C. acuminatus was 16,579 bp in length and had been deposited in GenBank with accession number of KX602324. In comparison with the other fish (Zhang et al. Citation2016), the mitogenome of C. acuminatus share the same organization consisting of 13 protein-coding genes(PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a putative control region. Except for ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Pro, and tRNA-Glu), which are encoded on the light strand (L-strand), the remaining genes are encoded on the heavy strand (H-strand). The overall nucleotide composition of C. acuminatus mitochondrial genome is A:34.05%, T:32.76%, G:13.36%, and C:19.83%, with the A + T content of 66.81%, showing an obvious anti-G bias in accordance with the mitochondrial genomes of other teleost species (Norfatimah et al. Citation2014; Xie et al. Citation2016). Among all 13 protein-coding genes, we found that most protein-coding genes for C. acuminatus share the common initiation codon ATG, while only COXI gene which start from GTG instead of ATG. Besides, incomplete termination codons (T or TA) were also found in six genes (ND2, COXII, COXIII, ND3, ND4, and Cytb), which may be completed by polyadenylation of the RNA messenger after cleavage (Nardi et al. Citation2001) ().
Table 1. Characteristics of the mitochondrial genome of C. acuminatus.
There are seven regions of gene overlap ranging from 1 to 7 bp and 13 intergenic spacer regions ranging from 1 to 35 bp with the longest intergenic region appeared between tRNA-Asn and tRNA-Cys. Overlapped gene was believed to be associated with the transition from RNA to DNA synthesis (Hixson et al. Citation1986) ().
Similarly to other mitochondrial genomes, the two ribosomal RNA genes (12S rRNA and 16S rRNA) are located between tRNA-Phe and tRNA-Leu within C. acuminatus mitogenome, and separated by tRNA-Val. Besides, our analysis indicated that 22 tRNA genes varying from 67 to 76 bp are interspersed throughout the genome. The control region (D-loop) is 928 bp in length which is located between the tRNA-Pro and tRNA-Phe genes, as generally shown in most vertebrate mitochondrial genome (Liu & Yang Citation2013; Quan et al. Citation2013) ().
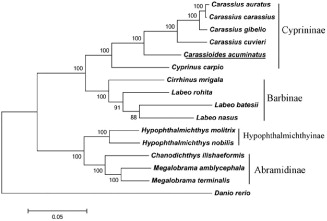
The phylogenetic tree was constructed on the basis of the complete mitogenome sequences from C. acuminatus and other 15 closely related species in the GenBank database. A neighbour-joining tree was constructed by using MEGA 5.1 Program. According to the established phylogenetic tree, we confirm that the C. acuminatus is much closer to Carassius, which coincides to the morphological taxonomy ().
Figure 1. The phylogenetic tree based on 16 related mitochondrial genomes. The accession numbers of the species are C. acuminatus (KX602324.1), Carassius auratus (AB111951.1), C. carassius (AY714387.1), Carassius cuvieri (AP011237.1), C. gibelio (JF496198.1), Chanodichthys ilishaeformis (NC_029722.1), Cirrhinus mrigala (JQ838173.1), Cyprinus carpio (X61010.1), Danio rerio (AC024175.3), Hypophthalmichthys molitrix (EU315941.1), H. nobilis (AP011217.1), Labeo batesii (AB238967.1), L. nasus (AP013333.1), L. rohita (AP011201.1), Megalobrama amblycephala (AP011219.1) and M. terminalis (AB626850.1).
Disclosure statement
The authors report that they have no conflicts of interest. The authors alone are responsible for the content and writing of the paper. This work was financially supported by The Public Sector (Agriculture) Special Scientific Research Projects (201303056-5).
References
- Chunxing D, Wenyi Z, Xingchen G, Zhongneng X, Qun Z. 2014. Genetic diversity of Carassioides cantonensis of Hainan island based on mitochondrial control region sequences analysis. Guangdong Agri Sci. 9:151–154. (Chinese)
- Hixson JE, Wong TW, Clayton DA. 1986. Both the conserved stem-loop and divergent 5′-flanking sequences are required for initiation at the human mitochondrial origin of light-strand DNA replication. J Biol Chem. 261:2384–2390.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Liu Y, Yang J. 2013. The complete mitochondrial genome sequence of Xingkai topmouth culter (Culter alburnus). Mitochondrial DNA. 25:451–453.
- Nardi F, Carapelli A, Fanciulli PP, Dallai R, Frati F. 2001. The complete mitochondrial DNA sequence of the basal hexapod tetrodontophora bielanensis: evidence for heteroplasmy and tRNA translocations. Mol Biol Evol. 18:1293–1304.
- Norfatimah MY, Teh LK, Salleh MZ, Isa MM, SitiAzizah MN. 2014. Complete mitochondrial genome of Malaysian Mahseer (Tor tambroides). Gene. 548:263–269.
- Quan XQ, Jin XX, Sun YN. 2013. The complete mitochondrial genome of Lophiogobius ocellicauda (Perciformes, Gobiidae). Mitochondrial DNA. 25:95–97.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Xie XY, Huang GF, Li YT, Zhang YT, Chen SX. 2016. Complete mitochondrial genome of Acrossocheilus parallens (Cypriniformes, Barbinae). Mitochondrial DNA Part A. 27:3339–3340.
- Zhang Z, Zhang N, Liu M, Gao T. 2016. The complete mitochondrial genome of Coilia grayii (Clupeiformes: Engraulidae). Mitochondrial DNA Part A. 27:3175–3176.
- Zheng C. 1989. The fishes of Pearl River [M]. Beijing: Science Press. (Chinese)
