Abstract
The rockweed F. distichus is a one of the most common intertidal seaweeds in the northern hemisphere. The systematics of F. distichus however remains open to discussion. Here, we contribute to the bioinformatics and systematics of F. distichus by deciphering its complete mitogenome. The F. distichus mitogenome is 36,400 bp in length, contains 67 genes, and has a gene content, organization, and sequence that are similar to the generitype, F. vesiculusus. These data support the continued recognition of F. distichus as a polymorphic entity with a broad distribution and high degree of ecological diversity.
Fucus L. is one of the three original genera proposed by Linnaeus (Citation1753) to accommodate macroscopic marine algae. Since then more than 1000 names have been assigned to the genus (Guiry & Guiry Citation2017), but nearly all have been reclassified. Based on genetic analyses, three or four species are recognized: F. distichus L., F. serratus L., F. spiralis L., and F. vesiculosus L. (Serrao et al. Citation1999; Coyer et al. Citation2006; Kucera & Saunders Citation2008; Coyer et al. Citation2011; Laughinghouse et al. Citation2015). This classification remains problematic however, as exemplified by F. distichus, which displays a high degree of morphological, ecological, geographical, and reproductive variation (Gardner Citation1922; Kucera & Saunders Citation2008; Coyer et al. Citation2011; Laughinghouse et al. Citation2015). Herein, we characterize the mitogenome of F. distichus to clarify its systematics.
Fucus distichus (Voucher- UC 2050487) was collected from Coos Bay, Oregon (43°21′30.3″, −124°18′32.2″) and its DNA isolated following Lindstrom et al. (Citation2011). The 76 bp paired-end library construction and sequencing was performed by myGenomics, LLC (Alpharetta, GA,) yielding 22,040,820 reads. The mitogenome was assembled using the default de novo settings in CLC Genomics Workbench 9.5 (®2016 CLC bio, a QIAGEN Company, Waltham, MA) and annotated using blastx and NCBI ORF-finder. The mitogenome was aligned to other Phaeophyceae with MAFFT (Katoh & Standley Citation2013). The RaxML analysis was executed using complete mitogenome sequences at Trex-online (Boc & Makarenkov Citation2012) with the GTR + gamma model and 1000 fast bootstraps, then visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
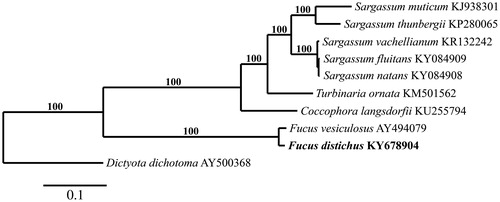
The F. distichus mitogenome (GenBank KY678904) is 36,400 bp in length and contains cob and tatC, 3 rRNA, 3 open reading frames, 3 ATP synthase, 3 cox, 6 rpl, 10 NADH, 11 rps, and 26 tRNA genes (trnL occurs in triplicate and trnI, trnM, trnS, trnY in duplicate). Gene content, organization, and length are nearly identical to F. vesiculosus (36,392 bp) (Oudot-Le Secq et al. Citation2006). The genetic distance between these two species is 2.0%. Phylogenetic analysis of F. distichus supports this close relationship, positioning it in a clade with F. vesiculosus (). Sequence analysis of F. distichus from Oregon identifies it as haplotype ic6 (Coyer et al. Citation2011) from Japan and Alaska, USA. Its cox1 sequence was also an exact match to a specimen from British Columbia, Canada (GenBank EU646633), and only differed by 1 bp from F. distichus from Nova Scotia, Canada (GenBank EU646647), Nordland, Norway (GenBank LN877838), and California, USA (GenBank KM254965).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Boc A, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40:W573–W579.
- Coyer JA, Hoarau G, Oudot-Le Secq MP, Stam WT, Olsen JL. 2006. A mtDNA-based phylogeny of the brown algal genus Fucus (Heterokontophyta; Phaeophyta). Mol Phylogenet Evol. 39:209–222.
- Coyer JA, Hoarau G, Van Schaik J, Luijckx P, Olsen JL. 2011. Trans‐Pacific and trans‐Arctic pathways of the intertidal macroalga Fucus distichus L. reveal multiple glacial refugia and colonizations from the North Pacific to the North Atlantic. J Biogeogr. 38:756–771.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469.
- Gardner NL. 1922. The genus Fucus on the Pacific coast of North America. Univ Calif Publ Bot. 10:1–180.
- Guiry MD, Guiry GM. 2017. AlgaeBase. [Internet]. Galway: World-wide electronic publication, National University of Ireland; [cited 2017 Feb 16]. Available from: http://www.algaebase.org
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kucera H, Saunders GW. 2008. Assigning morphological variants of Fucus (Fucales, Phaeophyceae) in Canadian waters to recognized species using DNA barcoding. Botany. 86:1065–1079.
- Laughinghouse HD, IV Müller KM, Adey WH, Lara Y, Young R, Johnson G. 2015. Evolution of the northern rockweed, Fucus distichus, in a regime of glacial cycling: implications for benthic algal phylogenetics. PLoS One. 10:e0143795.
- Lindstrom SC, Hughey JR, Martone PT. 2011. New, resurrected and redefined species of Mastocarpus (Phyllophoraceae, Rhodophyta) from the northeast Pacific. Phycologia. 50:661–683.
- Linnaeus C. 1753. Species plantarum, exhibentes plantas rite cognitas, ad genera relatas, cum differentiis specificis, nominibus trivialibus, synonymis selectis, locis natalibus, secundum systema sexuale digestas. Holmiae (Stockholm): Impensis Laurentii Salvii.
- Oudot-Le Secq MP, Loiseaux-de Goer S, Stam WT, Olsen JL. 2006. Complete mitochondrial genomes of the three brown algae (Heterokonta: Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr Genet. 49:47–58.
- Serrao EA, Alice LA, Brawley SH. 1999. Evolution of the Fucaceae (Phaeophyceae) inferred from nrDNA-ITS. J Phycol. 35:382–394.