Abstract
The complete mitochondrial genome of a treefrog (Hyla sp.) was determined. The circular mitochondrial genome is 18,288 bp long and encodes 13 proteins, 22 transfer RNAs, and 2 ribosomal RNAs. Phylogenetic analysis of its full genome sequences showed that H. sp. was closely related to H. ussuriensis and H. japonica rather than Dryophytes suweonensis, consistent with the results from each protein-coding gene and a cluster between 12S rRNA and 16S rRNA. The present study will provide essential genomic information for biogeographical distribution and evolutionary history of an endemic treefrog, H.sp.
Keywords:
The genus Hyla distributed widely in North America, Eurasia, and Africa, with more than 35 species (Li et al. Citation2015). Of these, two treefrog species such as Hyla japonica and H. suweonensis have been identified in Korea. Unlike H. japonica with a widespread distribution throughout North East Asia, the H. suweonensis is endemic to Korea, with a restricted distribution around the western coastal plains of Korea. Systematics and biogeography of the genus Hyla on the East Asian species indicates that H. japonica and H. suweonensis are potentially synonymous to H. ussuriensis and H. immaculate from China, respectively (Li et al. Citation2015). Divergence-time estimation, diversification and ancestral-area reconstruction analyses also suggest that genus Hyla can be changed to Dryophytes (Li et al. Citation2015; Duellman et al. Citation2016). However, the complete mitochondrial genome of D. suweonensis (as the synonym of an endangered treefrog, H. suweonensis) was not generated until recently (Borzée et al. Citation2017), and its genetic information on the geographical distribution is unfortunately limited. Here, we sequenced the complete mitochondrial genomes of an endemic treefrog, H. sp. by next-generation sequencing (NGS) technology, which can be helpful for its evolutionary and phylogenetic analyses.
The specimens of H. sp. for buccal swabbing were collected from Iksan (35.950° N; 126.958° E), Korea, and the total genomic DNA was extracted from an adult male (stored at the Wildlife Specimen Bank in Chonnam National University, Korea), considering initially as a H. suweonensis based on its vocal characteristics, habitat selection, and morphological properties (Ham Citation2014). Following quality control and library construction of the specimen, the NGS reads (450-bp length in each read) generated from MiSeq (Macrogen, Seoul, Korea). Approximately 0.25% mapped reads (71,863 out of 28,476,666) were used for de novo assembly and annotation by using commercial software (MITOS) to identify a complete mitochondrial genome with about an average 150 × coverage.
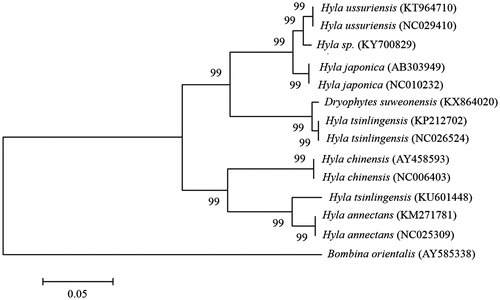
The complete mitochondrial genome of H. sp. was 18,288 bp in length, which contains 37 genes for 13 proteins, 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), an origin of light strand replication site (OL), and a long noncoding region called the control or D-loop region with a 202bp gap (GenBank: KY700829). The overall nucleotide composition for H. sp. was 30.1% A, 25.5% C, 14.3% G, and 30.1% T. The phylogenetic analysis was performed using MEGA6 to construct a neighbour-joining tree with 1000 bootstrap replicates containing complete mitochondrial genomes of 13 species derived from sister-genera Hyla and Dryophytes (). Bombina orientalis was used as an outgroup for tree rooting. The phylogenetic tree indicated that H. sp. showed closer evolutionary distances with H. ussuriensis compared with D. suweonensis (Borzée et al. Citation2017). Variants from the full genome sequences of H. sp. were approximately 10% when compared to that of D. suweonensis, although the specimen used in the present study was considered initially to D. suweonensis based on its morphological and vocal features (Ham Citation2014). The variation in amino acid sequences from 13 protein coding genes was an average of 0.90% (range: 0–2.65%) between H. sp. and H. japonica, and 3.06% (0.19–5.67%) between H. sp. and D. suweonensis. The phylogenetic relationships based on amino acid sequences of each protein-coding gene and a cluster between 12S rRNA and 16S rRNA (nucleotides of 12S rRNA – tRNAVal – 16S rRNA) also supports the aforementioned phylogenetic relationships among the genera Hyla and Dryophytes species ().
Figure 1. Phylogeny of H. sp and other related species based on complete mitochondrial genome sequences. The complete mitochondrial genomes were downloaded from GenBank and the phylogenetic tree is constructed by a neighbour-joining method with 1000 bootstrap replicates containing the full genomes of 13 species derived from Hyla and Dryophytes. B. orientalis was used as an outgroup for tree rooting. GenBank accession numbers of each mitochondrial genome sequence are given in the bracket after the species name.
The complete mitochondrial genome sequences of H. sp. may provide intriguing insight into biogeographical distribution and conservation genetics of this endemic species, and to evoke to do further researches to reveal the behavioural and morphological relationships among Hyla and Dryophytes species.
Disclosure statement
The authors declare no conflict of interests.
References
- Borzée A, Didinger C, Jang Y. 2017. Complete mitochondrial genome of Dryophytes suweonensis (Anura Hylidae). Mitochondrial DNA Part B. 2:5–6.
- Duellman WE, Marion AB, Hedges SB. 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa. 4104:1–109.
- Ham CH. 2014. Morphology, age structure and mating call characteristics of Japanese tree frog (Hyla japonica) and Suweon tree frog (Hyla suweonensis) [thesis]. Gwangju, South Korea: Chonnam National University.
- Li J-T, Wang J-S, Nian H-H, Litvinchuk SN, Wang J, Li Y, Rao D-Q, Klaus S. 2015. Amphibians crossing the Bering Land Bridge: Evidence from Holarctic treefrogs (Hyla, Hylidae, Anura). Mol Phylogenet Evol. 87:80–90.
