Abstract
The complete mitochondrial genome of the burrowing ghost shrimp Nihonotrypaea harmandi was reconstructed using the Illumina HiSeq platform. The genome was 15,272 bp in length made up of 37 mitochondrial genes (13 CDSs, 22 tRNAs, and 2 rRNAs) in the same order as other Nihonotrypaea species. Phylogenetic analyses suggested that Nihonotrypaea is a valid genus, and that N. harmandi can be phylogenetically marginally separated from N. japonica, though some authors considered the former as a synonym of the latter.
The ghost and mud shrimps (former infraorder Thalassinidea, cf. Poore et al. Citation2014) play an important role as ecosystem engineers and affect ecosystem processes and community structure (Pillay & Branch Citation2011). The genus Nihonotrypaea (Manning & Tamaki Citation1998) is a callianassid ghost shrimp, and distinguished from other genera of the same subfamily, Callianassinae, by the appendices internae projecting from the margin of the endopod of pleopods 3–5 (Manning & Tamaki Citation1998). The status of Nihonotrypaea is, however, controversial, being regarded as a synonym of Trypaea Dana, 1852 (Sakai Citation2011). Currently, six species of Nihonotrypaea are described from the Northwest Pacific, of which three species, N. harmandi (Bouvier Citation1901), N. japonica (Ortmann Citation1891), N. petalura (Stimpson Citation1860), are distributed in an estuarine system in mid-western Kyushu, southern Japan, each inhabiting different environmental conditions (Tamaki et al. Citation1999), whereas Sakai (Citation2011) considered N. harmandi and N. japonica belonging to one single species, Nihonotrypaea (= Trypaea) japonica. Here we determined the complete mitochondrial genome of N. harmandi, which was phylogenetically analyzed in order to clarify the status of the genus Nihonotrypaea and the relationships between N. harmandi and N. japonica.
An approximate of 30 mg of a male’s major cheliped-muscle was dissected from a live specimen of N. harmandi (Specimen Voucher: Nagasaki University #Call160314) collected from an intertidal sandflat in Koyagi, Nagasaki (129°47.4′E, 32°41.4′N) on 22 January 2016. Total DNA was extracted, and whole genome sequencing was outsourced to Macrogen (Seoul, South Korea). A total of 65M 101-bp paired-end reads generated by Illumina HiSeq 4000 were assembled using IDBA_UD (Peng et al. Citation2012). A circular contig which agreed with the known Nihonotrypaea mitochondrial genomes was annotated with MITOS (Bernt et al. Citation2012) followed by manual validation of the coding regions using the reference genomes. Phylogenetic analyses were conducted using MEGA7 (Kumar et al. Citation2016).
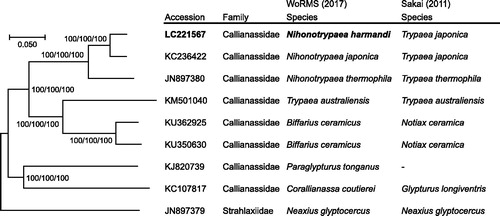
The complete mitochondrial genome of N. harmandi was 15,272 bp in length (GenBank accession number: LC221567), and contained 37 mitochondrial genes (13 CDSs, 22 tRNAs, and 2 rRNAs) in the same order as N. japonica (accession number: KC236422) and N. thermophile (accession number: JN897380). A phylogenetic tree reconstructed from a combined analysis of 13 CDSs and 2 rRNAs agreed with the classification of genera by WoRMS Editorial Board (Citation2017) (). Furthermore, despite the considerable morphological similarities between the species of Nihonotrypaea and Biffarius (cf. Liu & Liu Citation2014), the two genera formed distinct monophyletic clades that are well-separated from each other, suggesting the validity of the genus Nihonotrypaea. The two species, N. harmandi and N. japonica, were closely related with each other, while a Kimura-2-parameter (K2P) distance value for COI gene nucleotide sequence, which can be an index for species delimitation (e.g. Hebert et al. Citation2003), was 6.67% between these species. Given that the maximum intraspecific K2P value is shown to be no more than 5% in Decapoda (Costa et al. Citation2007; Matzen da Silva et al. Citation2011; Raupach et al. Citation2015), N. harmandi can be phylogenetically marginally separated as a species from N. japonica.
Figure 1. Phylogenetic relationships of the callianassid ghost shrimps inferred from a combined analysis of 13 CDSs and 2 rRNAs using NJ (K2P model), ML (GTR + I + G model) and MP methods each with 1,000 replicates of bootstrap. There were a total of 12,675 positions in the dataset. The tree shown is an NJ tree, and the ML and MP trees were the same topology. Numbers above nodes are bootstrap support values (NJ/ML/MP). Neaxius glyptocercus (accession number: JN897379) was used as outgroup. Classifications of species proposed by WoRMS Editorial Board (Citation2017) and Sakai (Citation2011) are listed. The original species name (source organism in GenBank) of the sequences used for tree reconstructions are listed as follows: KC236422 (Nihonotrypaea japonica), JN897380 (Nihonotrypaea thermophile), KM501040 (Trypaea australiensis), KU362925 (Callianassa ceramica), KU350630 (Callianassa ceramica), KJ820739 (Paraglypturus tonganus), KC107817 (Corallianassa coutierei), JN897379 (Neaxius glyptocercus).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2012. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bouvier EL. 1901. Sur quelques crustacés du Japon, offerts au muséum par M. le Dr. Harmand. Bull Mus Hist nat, Paris. 7:332–334.
- Costa FO, deWaard JR, Boutillier J, Ratnasingham S, Dooh RT, Hajibabaei M, Hebert PDN. 2007. Biological identifications through DNA barcodes: the case of the Crustacea. Can J Fish Aquat Sci. 64:272–295.
- Hebert PDN, Ratnasingham S, deWaard JR. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 270 Suppl 1:S96–S99.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu W, Liu R. 2014. A new species of the genus Nihonotrypaea Manning & Tamaki, 1998, Nihonotrypaea hainanensis sp. n., collected from the South China Sea. ZooKeys. 457:35–44.
- Manning RB, Tamaki A. 1998. A new genus of ghost shrimp from Japan (Crustacea: Decapoda: Callianassidae). Proc Biol Soc Washington. 111:889–892.
- Matzen da Silva J, Creer S, dos Santos A, Costa AC, Cunha MR, Costa FO, Carvalho GR. 2011. Systematic and evolutionary insights derived from mtDNA COI barcode diversity in the Decapoda (Crustacea: Malacostraca). PLoS One. 6:e19449.
- Ortmann AE. 1891. Die Decapoden-Krebse des Straßburger Museums, mit besonderer Berücksichtigung der von Herrn Dr. Döderlein bei Japan und bei den Liu-Kiu-Inseln gesammelten und z. Z. im Straßburger Museum aufbewahrten Formen. III. Die Abtheilungen der Reptantia Boas: Homaridae, Loricata und Thalassinidea. Zool Jb (Systematik, Geographie und Biologie der Thiere). 6:1–58.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28:1420–1428.
- Pillay D, Branch GM. 2011. Bioengineering effects of burrowing thalassinidean shrimps on marine soft-bottom ecosystems. Oceanogr Mar Biol Ann Rev. 49:137–192.
- Poore GCB, Ahyong ST, Bracken-Grissom HD, Chan T-Y, Chu KH, Crandall KA, Dworschak PC, Felder DL, Feldmann RM, Hyžný M, et al. 2014. On stabilising the names of the infraorders of thalassinidean shrimps, Axiidea de Saint Laurent, 1979 and Gebiidea de Saint Laurent, 1979 (Decapoda). Crustaceana. 87:1258–1272.
- Raupach MJ, Barco A, Steinke D, Beermann J, Laakmann S, Mohrbeck I, Neumann H, Kihara TC, Pointer K, Radulovici A, et al. 2015. The application of DNA barcodes for the identification of marine crustaceans from the North Sea and adjacent regions. PLoS One. 10:e0139421.
- Sakai K. 2011. Axioidea of the world and a reconsideration of the Callianassoidea (Decapoda, Thalassinidea, Callianassida). Crustaceana Monographs. 13:1–616.
- Stimpson W. 1860. Prodromus descriptionis animalium evertebratorum, quae in expeditione ad Oceanum Pacificum septentrionalem, a Republica Federata missa, Cadwaladaro Ringgold et Johanne Rodgers ducibus, observavit et descripsit. Pars VIII. Crustacea Macrura. Proc Acad nat Sci Philadelphia. 1860:22–47.
- Tamaki A, Itoh J, Kubo K. 1999. Distributions of three species of Nihonotrypaea (Decapoda. Thalassinidea: Callianassidae) in intertidal habitats along an estuary to open-sea gradient in western Kyushu, Japan. Crust Res. 28:37–51.
- WoRMS Editorial Board. 2017. World register of marine species; [cited 2017 Mar 03]. Available from: http://www.marinespecies.org at VLIZ
