Abstract
The plant genus Melastoma is comprised of members estimated to have formed through recent species radiation. Natural hybridization among member species further complicates taxonomy within the genus. Herein, we report the complete chloroplast genome of M. candidum, assembled from partial data obtained from a parallel whole-genome Illumina paired-end sequencing effort on the species. The chloroplast genome was 156,682 bp in length, with a large single-copy (LSC) region of 86,084 bp, a small single-copy (SSC) region of 17,094 bp, separated by two inverted repeat (IR) regions of 26,752 bp each. It was predicted to contain a total of 129 genes, with an overall GC content of 37.17%. Phylogenetic analysis placed M. candidum in the same clade as species within the Melastomeae tribe of Melastomataceae.
The plant genus Melastoma comprises of species distributed mainly in the tropical and subtropical regions of Asia and Oceania (Meyer Citation2001). The recognized number of species remains debatable until today, with claims ranging from 22 (Meyer Citation2001), 80–90 (Wong Citation2016), to about 100 (Chen Citation1984) species. The diversity observed in Melastoma is thought to be the outcome of species radiation, estimated to have happened within the past one million years (Renner & Meyer Citation2001). Recent studies have also found widespread hybridization among the different species (e.g. Dai et al. Citation2012; Liu et al. Citation2014; Wong Citation2015), further complicating species delineation within the genus. Here, we report the complete chloroplast genome sequence of M. candidum, commonly found in southern China, as a resource for future studies on the taxonomy of Melastoma.
Sequence data used for the assembly of this chloroplast genome was extracted from the total sequencing data from a parallel whole-genome Illumina paired-end sequencing effort of an M. candidum individual (Wu et al., unpublished data) sampled from Wenchang, Hainan, China. The voucher specimen (MCAN-2013-HN01) is kept at the Sun Yat-sen University Herbarium (SYS). Approximately 4 Gb of paired-end (125 bp) sequence data was randomly extracted from the total sequencing output, as input into NOVOPlasty (Dierckxsens et al. Citation2017) to assemble the chloroplast genome. A partial chloroplast rbcL gene sequence of the same species (GenBank accession GQ436728) was used as the seed sequence for the seed-and-extend algorithm implemented in NOVOPlasty. The accuracy of the automated assembly was then verified by Sanger-sequencing of seven randomly chosen genes (rps16, atpF, rpoC2, rpoC1, atpE, rpoA, and ndhD), covering approximately 14 kbp (∼8.9% of the total chloroplast genome size), with 100% accuracy. Annotation of the chloroplast genome was performed using Verdant (McKain et al. Citation2017) and DOGMA (Wyman et al. Citation2004), then manually verified and corrected by comparison with sequences on GenBank.
The complete chloroplast genome sequence of M. candidum (GenBank accession KY745894) obtained in this study was 156,682 bp in length, with a large single-copy (LSC) region of 86,084 bp, a small single-copy (SSC) region of 17,094 bp, separated by two inverted repeat (IR) regions of 26,752 bp each. It was predicted to contain 129 genes, including 85 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. The overall GC content was 37.17%. The ndhD gene had an ACG start codon, instead of the conventional AUG start codon.
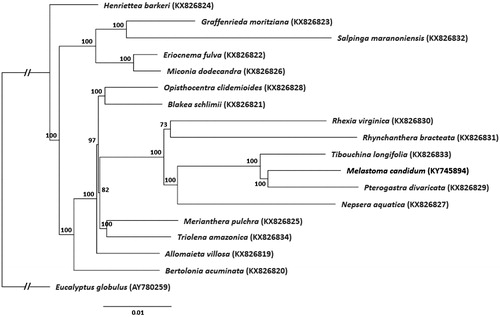
For phylogenetic tree construction, the chloroplast genome of M. candidum was aligned with 16 other complete chloroplast genome sequences of Melastomataceae (Reginato et al., Citation2016) and Eucalyptus globulus (AY780259) as outgroup, using MAFFT v7.307 (Katoh & Standley Citation2013). A maximum likelihood tree () was then constructed using RAxML (Stamatakis, Citation2014). Melastoma candidum shared the same clade with two other genera within the Melastomeae tribe, Petrogastra and Tibouchina.
Acknowledgements
We thank Nicolas Dierckxsens for the assistance in using NOVOPlasty, and Michael McKain for the assistance in using Verdant.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chen J. 1984. Melastomataceae. In: Wu ZY, Raven PH, editors. 2007. Flora of China. Vol. 13. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. p. 360–399.
- Dai S, Wu W, Zhang R, Liu T, Chen Y, Shi S, Zhou R. 2012. Molecular evidence for hybrid origin of Melastoma intermedium. Bioch Syst Ecol. 41:136–141.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu T, Chen Y, Chao L, Wang S, Wu W, Dai S, Wang F, Fan Q, Zhou R. 2014. Extensive hybridization and introgression between Melastoma candidum and M. sanguineum. PLoS One. 9:e96680.
- McKain MR, Hartsock RH, Wohl MM, Kellogg EA. 2017. Verdant: automated annotation, alignment and phylogenetic analysis of whole chloroplast genomes. Bioinformatics. 33:130–132.
- Meyer K. 2001. Revision of Southeast Asian genus Melastoma (Melastomataceae). Blumea. 46:351–398.
- Reginato M, Neubig KM, Majure LC, Michelangeli FA. 2016. The first complete plastid genomes of Melastomataceae are highly structurally conserved. PeerJ. 4:e2715.
- Renner SS, Meyer K. 2001. Melastomeae come full circle: biogeographic reconstruction and molecular clock dating. Evolution. 55:1315–1324.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wong KM. 2015. Studies in Southeast Asian Melastoma (Melastomataceae), 1. Morphological variation in Melastoma malabathricum and notes on rheophytic taxa and interspecific hybridisation in the genus. Gardens’ Bulletin Singapore. 67:387–401.
- Wong KM. 2016. The Genus Melastoma in Borneo including 31 new species. Kota Kinabalu: Natural History Publications; Singapore: National Parks Board.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.