Abstract
The complete mitochondrial genome was sequenced in two individuals of the Sakhalin sculpin Cottus amblystomopsis. The genome sequences are 16,526 and 16,527 bp in size, and the gene arrangement, composition, and size are very similar to the other sculpin mitochondrial genomes published previously. The difference between the two genomes studied is low, 0.28%, in spite of the relatively long distance separating the localities. The data are consistent with the amphidromous life history of C. amblystomopsis, promoting gene flow even between distantly located rivers.
The Sakhalin sculpin Cottus amblystomopsis Schmidt is an amphidromous fish distributed widely in the rivers of the Sea of Japan and the Sea of Okhotsk including Hokkaido Island, Southern Sakhalin Island, and Southern Kuril Islands (Shedko Citation2001; Nakabo Citation2002; Chereshnev Citation2003). Adult sculpins inhabit and spawn in freshwater and their larvae drift downstream to marine environments where they spend about one month, after which they return back to rivers as juveniles (Goto Citation1990). Kinziger et al. (Citation2005) treated the Sakhalin sculpin C. amblystomopsis (along with C. nozawae, endemic to the Japan Islands) as a Cephalocottus clade or subgenus within the family Cottidae. Goto et al. (Citation2015) considered C. amblystomopsis within the genus Cottus as the lineage C from East Asia. The evolutionary history and phylogenetic relationships of C. amblystomopsis remained unclear due to limited genetic data available for the species.
We have sequenced two complete mitochondrial (mt) genomes of C. amblystomopsis (GenBank accession numbers KY563345 and KY563346) from the Taranai River (46°37.76′ N; 142°24.31′ E), Sakhalin Island, and the Djigitovka River (44°50.06′ N; 136°06.95′ E), Primorsky krai, Russia, using primers designed with the program mitoPrimer_V1 (Yang et al. Citation2011). The fish specimens are stored at the museum of the A. V. Zhirmunsky Institute of Marine Biology, National Scientific Center of Marine Biology, Vladivostok, Russia (www.museumimb.ru) under accession numbers MIMB 33262 and MIMB 33263. The genome sequences are 16,526 and 16,527 bp in size and the gene arrangement, composition, and size are very similar to the sculpin fish genomes published previously. We detected 47 single nucleotide and one length differences between the haplotypes CAM1-13 and CAM1-14; total sequence divergence (Dxy) is 0.0028 ± 0.0005.
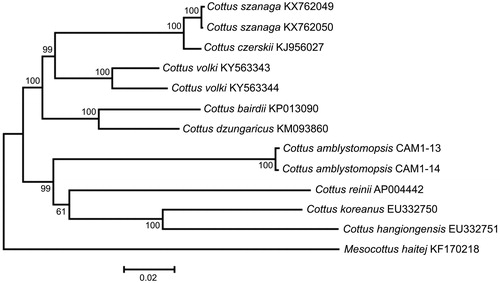
Comparison of the two mt genomes now obtained with other complete mt genomes available in GenBank for the genera Cottus and Mesocottus reveals a close affinity of C. amblystomopsis to the cluster C. reinii + C. hangiongensis+ C. koreanus within the genus Cottus (). The difference between the two C. amblystomopsis mt genomes now studied is low, 0.28%, in spite of relatively long distance separating the localities (∼1670 km along the coast line). The data are consistent with the amphidromous life history of C. amblystomopsis (Goto Citation1990), promoting gene flow between distantly located rivers (Goto & Andoh Citation1990). The juveniles returning to freshwater environment are suggested to derive from larval pools originating from multiple source rivers, where larval mixing and long-range dispersal result in high connectivity across the coasts of Sakhalin Island and Primorye Territory. Thus, the amphidromous life history of C. amblystomopsis might constraint intraspecific divergence as was also shown for other amphidromous fishes (Chubb et al. Citation1998).
Figure 1. Maximum likelihood tree for the Sakhalin sculpin Cottus amblystomopsis specimens CAM1-13 and CAM1-14, and GenBank representatives of the family Cottidae. The tree is constructed using whole mt genome sequences. The tree is based on the General Time Reversible + gamma + invariant sites (GTR + G + I) model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Acknowledgements
We greatly appreciate Dr. S. S. Makeev (Yuzhno-Sakhalinsk, Russia) for the Cottus amblystomopsis specimen from the Sakhalin Island.
Disclosure statement
The funders had no role in study design, data collection, data analysis, decision to publish, or preparation of the manuscript. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chereshnev IA. 2003. New data on morphology and biology of the poor studied sculpins genus Cottus (Cottidae, Scorpaeniformes) of the Kunashir island. In: Ya V, editor. Levanidov’s biennial memorial meetings. Vladivostok: Dalnauka; p. 368–376.
- Chubb AL, Zink RM, Fitzsimons JM. 1998. Patterns of mtDNA variation in Hawaiian freshwater fishes: the phylogeographic consequences of amphidromy. J Hered. 89:8–16.
- Goto A. 1990. Alternative life-history styles in the Japanese freshwater sculpins revisited. Env Biol Fish. 28:101–112.
- Goto A, Andoh T. 1990. Genetic divergence between the sibling species of river-sculpin, Cottus amblystomopsis and C. nozawae, with special reference to speciation. Env Biol Fish. 28:257–266.
- Goto A, Yokoyama R, Sideleva VG. 2015. Evolutionary diversification in freshwater sculpins (Cottoidea): a review of two major adaptive radiations. Env Biol Fish. 98:307–335.
- Kinziger AP, Wood RM, Neely DA. 2005. Molecular systematics of the genus Cottus (Scorpaeniformes: Cottidae). Copeia. 2005:303–311.
- Nakabo T. 2002. Cottidae. In: Nakabo T, editor. Fishes of Japan with pictorial keys to the species. Volume 1. Tokyo: Tokai University Press; p. 628–650.
- Shedko SV. 2001. A list of Cyclostomata and freshwater fish from the coast of Primorye. In: Ya V, editor. Levanidov’s Biennial Memorial Meetings. Issue 1. Vladivostok: Dalnauka; p. 229–249.
- Yang CH, Chang HW, Ho CH, Chou YC, Chuang LY. 2011. Conserved PCR primer set designing for closely-related species to complete mitochondrial genome sequencing using a sliding window-based PSO algorithm. PLoS One. 6:e17729.
