Abstract
The complete mitochondrial genome of the western honey bee subspecies Apis mellifera meda was sequenced. This mitochondrial genome is 16,248 bp in length, with 37 classical eukaryotic mitochondrial genes and an A + T-rich region. Gene direction and arrangement are similar to those of other Apis mitogenomes. All genes initiate with ATT (six genes), ATG (four genes), ATA (two genes), and ATC (one gene) start codons and terminate with a TAA stop codon. Four genes are encoded on the heavy and nine on the light strands, respectively. All of the 22 tRNA genes, ranging from 66 to 78 bp, have a typical cloverleaf structure. The complete mitogenome of A.m. meda provides information on the biogeography and evolution of A. mellifera subspecies.
Apis mellifera meda is a subspecies of western honey bee native to Iran, northern Iraq, and southeastern Turkey (Sheppard Citation2015). Apis mellifera meda shares similarities with A.m. ligustica, A.m. anatolica, and honey bees in northern Iraq (Rahimi & Asadi Citation2010) and is similar, morphometrically, to A.m. ligustica, the Italian honey bee (Sheppard Citation2015). The complete mitochondrial genome of A.m. meda has not been studied. In this study, an adult honey bee worker of A.m. meda was obtained from the Ruttner Bee Collection at the Bee Research Institute at Oberursel, Germany (Voucher No. 3284, Pour-Elmi, Iran, 2004, 27°13N, 60°40E) and its mitogenome is reported (GenBank accession No. KY464957). The identity of the bee was confirmed morphometrically by the Institute staff. We followed the method of Eimanifar et al. (Citation2016) to extract and quantify genomic DNA. A genomic library was constructed from the genomic DNA using a Kapa Hyper Prep Kit (Kapa Biosystems, Woburn, MA) with a paired-end read (2 × 150) followed by next-generation sequencing on the Illumina Hi-Seq 3000/4000 (San Diego, CA). The mapping, assembly and annotation were performed as described previously (Eimanifar et al. Citation2017).
The complete sequence of A.m. meda is 16,248 bp in length and consists of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 putative control region (CR). The overall base composition of the A.m. meda mitogenome was 43.1% A, 41.7% T, 9.6% C, and 5.6% G. The gene content, structure, and arrangement of the A.m. meda mitogenome are similar to those observed in other Apis mitogenomes (Eimanifar et al. Citation2016), with four PCGs on the heavy strand with the remainder on the light strand. ATP6 and ATP8 share 19 nucleotides, though other PCGs do not overlap. Start codons included ATT (six PCGs), ATG (four PCGs), ATA (two PCGs), and ATC (one PCG), with all ending in a TAA stop codon.
The 16S rRNA and 12S rRNA were 1362 and 785 bp long, respectively, with 16S having a slightly higher AT content (84.4% versus 81.5%). These are located between the tRNA-Leu gene and CR, and were separated by the tRNA-Val gene. The 22 tRNAs all fold into typical a cloverleaf shape as identified by tRNAscan-SE (Lowe & Eddy Citation1997), and ranged in length from 66 to 78 bp. The 763 bp CR was 96.2% AT.
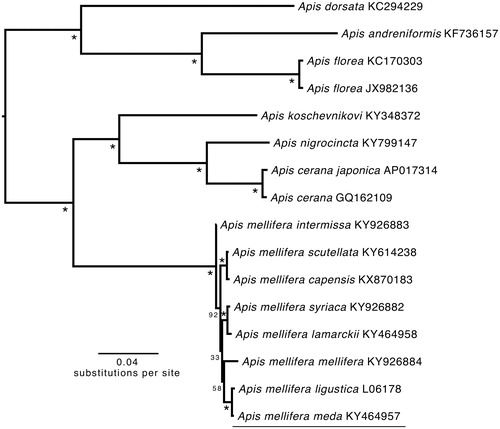
The phylogenetic position of A.m. meda was estimated from a concatenated dataset including the 13 PCGs and 2 rRNAs using the software RAxML 8.2.0 on the CIPRES science gateway (Miller et al. Citation2010) with GTRGAMMA and 1000 bootstrap replicates (). In the future, it is necessary to sequence the mitochondrial genomes of other A. mellifera subspecies to determine the phylogenetic relationships between them (especially the Middle Eastern subspecies) and A.m. meda. The p-distance between A.m. meda and A.m. liguistica is 0.004, while it ranged from 0.013 to 0.014 with A.m. mellifera individual. Additional mitogenomes from unstudied Apis subspecies will promote our understanding of the mitogenome evolution and diversity in Apis.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Eimanifar A, Kimball RT, Braun EL, Ellis JD. 2016. The complete mitochondrial genome of the Cape honey bee, Apis mellifera capensis Esch. (Insecta: hymenoptera: apidae). Mitochondrial DNA Part B. 1:817–819.
- Eimanifar A, Kimball RT, Braun EL, Ellis JD. 2017. The complete mitochondrial genome and phylogenetic placement of Apis nigrocincta Smith (Insecta: Hymenoptera: Apidae), an Asian, cavity-nesting honey bee. Mitochondrial DNA Part B. 2:249–250.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans (LA). 1–8.
- Rahimi A, Asadi M. 2010. Morphological characteristics of Apis mellifera meda (Hymenoptera: Apidae) in Saghez (west of Iran). Nature Montenegrina. 10:101–107.
- Sheppard WS. 2015. Honey bee diversity – races, ecotypes, strains. In: J. Graham, editor. The hive and the honey bee. Hamilton (IL): Dadant and Sons; p. 53–70.