Abstract
The greater green snake Cyclophiops major is a protected and colubrid species. Here, we investigated the complete mitochondrial genome of C. major. The genome is 17,217bp in size, including 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, 2 control regions, and an origin of light-strand replication. All genes are distributed on the heavy strand, except for ND6 gene and 8 tRNA genes. The AT content of the overall base composition of light strand is 59.83%, showing AT bias. Phylogenetic tree was built based on the genome of C. major and other related snakes to analyze their phylogenic relationship.
The greater green snake Cyclophiops major is a protected and docile species, and is considered non-venomous, belonging to the genus Cyclophiops of family Colubridae, which is widely distributed in China, Vietnam, and eastern Lao PDR (Orlov et al. Citation2000; Dieckmann et al. Citation2014; Ziegler et al. Citation2014). In this study, we determined the complete mitochondrial genome (mitogenome) of C. major (The GenBank accession number: KF148620). Samples of C. major (RE03064) were collected from Huang mount, Anhui Province of China, and deposited in the laboratory of the College of Life Sciences of Anhui Normal University, Wuhu, China. The total length of C. major is 17,217 bp. It consists of 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, 2 control regions (CRs, CRI and CRII), and an origin of light-strand replication (OL), which is similar to other reported alethinophidian snakes (He et al. Citation2010; Jang & Hwang, Citation2011; Li et al. Citation2016; Qian et al. Citation2016). The overall base composition of the light strand is as follows: A (34.61%), T (25.22%), C (27.89%), and G (12.28%), the AT content (59.83%) is significantly higher than the GC content (40.17%), which shows AT bias.
Except for ND6 gene and 8 tRNA genes, other genes are encoded by the heavy strand. Most PCGs use ATG as the start codon, while ND1, ND2, ND3 and COI genes use ATA, ATC, ATT and GTG as the start codon, respectively. Seven PCGs were terminated with the complete stop codon, ATP6, ATP8, ND4 and ND4L ended with TAA, COI gene stops with AGG, ND5 and ND6 using TAG as the stop codon, whereas other 6 PCGs ended with incomplete stop codon with T.
The length of the 12S rRNA gene is 923 bp, and is located between tRNAPhe and tRNAVal gene. 16S rRNA gene is 1478 bp, and is located between tRNAVal and ND1. There are 22 tRNA genes in C. major mitogenome, with the size ranging from 57 bp in tRNASer(AGY) to 73bp in tRNALeu(UUR). The sequence length of the CRI and CRII is 1050 and 1049 bp, respectively. The CRI is surrounded by tRNAPr° and tRNAPhe, while the CRII is located between tRNA Ile and tRNALeu(UUR). The OL is located between tRNAAsn and tRNACys gene, with a length of 36 bp in the WANCY cluster.
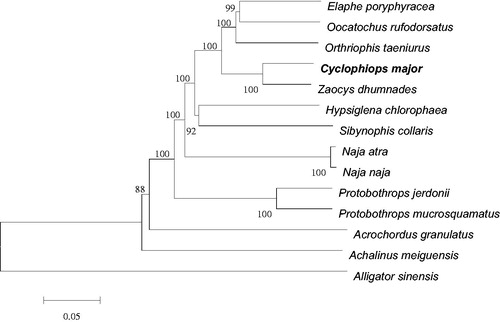
We used Mega6 (http://megasoftware.net) to build the neighbor-joining phylogenetic tree (), including mitogenome of C. major and other closely related 12 species that are from the family Colubridae, Acrochordidae, Elapidae, and Viperidae, which belong to Serpentes. Alligator sinensis was set as an outgroup. The results showed that C. major was clustered with the Z. dhumnades, highly supported by a bootstrap value of 100, and then clustered with O. taeniurus, O. rufodorsatus, E. Poryphyracea, H. Chlorophaea, S. Collaris, which supported the data that mitogenome of C. major are more closely related to other species of Colubridae family. The present study will provide a foundation for further research on the population genetics and systematic analyses of C. major, and provide genomic resources for Colubridae studies.
Figure 1. Neighbor-joining phylogenetic tree of complete mitogenome of C. Major and other related 12 species was constructed, with Alligator sinensis as an outgroup. The numbers on the each node branches are bootstrap values. All species’ accession numbers are listed as follows: Cyclophiops major KF148620; Zaocys dhumnades KF148621; Oocatochus rufodorsatus KC990020; Orthriophis taeniurus KC990021; Achalinus meiguensis NC_011576; Elaphe poryphyracea NC_012770; Hypsiglena chlorophaea NC_013977; Sibynophis collaris NC_016424; Acrochordus granulatus NC_007400; Naja atra NC_011389; Naja naja NC_010225; Protobothrops jerdonii NC_021402; Protobothrops mucrosquamatus NC_021412; Alligator sinensis NC_004448.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Dieckmann S, Norval G, Mao JJ. 2014. Notes on the reproductive biology of the greater green snake, Cyclophiops major (Günther, 1858), in Taiwan. IRCF Reptiles Amphib. 21:100–102.
- He M, Feng JC, Zhao EM. 2010. The complete mitochondrial genome of the Sichuan hot-spring kell-back (Thermophis zhaoermii; Serpentes: Colubridae) and amitogenomic phylogeny of the snakes. Mitochondrial DNA. 21:8–18.
- Jang KH, Hwang UW. 2011. Complete mitochondrial genome of the black-headed snake Sibynophis collaris (Squamata, Serpentes, Colubridae). Mitochondrial DNA. 22:77–79.
- Li E, Sun FX, Zhang RD, Chen J, Wu XB. 2016. The complete mitochondrial genome of the striped-tailed rat-snake, Orthriophis taeniurus (Reptilia, Serpentes, Colubridae). Mitochondrial DNA. 27:599–600.
- Orlov NL, Murphy RW, Papenfuss TJ. 2000. List of snakes of Tam-Dao mountain ridge (Tonkin, Vietnam). Russ J Herpetol. 7:69–80.
- Qian LF, Zhang CL, Huang X, Pan T, Wang H, Zhang BW. 2016. Mitochondrial genome of Dinodon rufozonatum (Squamata: Colubridae: Dinodon). Mitochondrial DNA. 27:970–971.
- Ziegler T, Tran DTA, Nguyen TQ, Perl RGB, Wirk L, Kulisch M, Lehmann T, Rauhaus A, Nguyen TT, Le QK, Vu TN. 2014. New amphibian and reptile records from Ha Giang Province, northern Vietnam. Herpetol Notes. 7:185–201.
